7EAL
 
 | |
5W4D
 
 | | C. japonica N-domain, Selenomethionine mutant | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CHLORIDE ION, ... | | Authors: | Aoki, S.T, Bingman, C.A, Kimble, J. | | Deposit date: | 2017-06-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | C. elegans germ granules require both assembly and localized regulators for mRNA repression.
Nat Commun, 12, 2021
|
|
1YIQ
 
 | | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADHIIG from Pseudomonas putida HK5. Compariison to the other quinohemoprotein alcohol dehydrogenase ADHIIB found in the same microorganism. | | Descriptor: | CALCIUM ION, HEME C, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Toyama, H, Chen, Z.W, Fukumoto, M, Adachi, O, Matsushita, K, Mathews, F.S. | | Deposit date: | 2005-01-12 | | Release date: | 2005-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADH-IIG from Pseudomonas putida HK5
J.Mol.Biol., 352, 2005
|
|
6PW0
 
 | | Cytochrome C oxidase delta 6 mutant | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, S. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural changes at the surface of cytochrome c oxidase alter the proton-pumping stoichiometry.
Biochim Biophys Acta Bioenerg, 1861, 2019
|
|
3HUD
 
 | | THE STRUCTURE OF HUMAN BETA 1 BETA 1 ALCOHOL DEHYDROGENASE: CATALYTIC EFFECTS OF NON-ACTIVE-SITE SUBSTITUTIONS | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Hurley, T.D, Bosron, W.F, Hamilton, J.A, Amzel, L.M. | | Deposit date: | 1993-01-04 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of human beta 1 beta 1 alcohol dehydrogenase: catalytic effects of non-active-site substitutions.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
7L0C
 
 | | Ligand-free PTP1B T177G | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Shen, R.D, Hengge, A.C, Johnson, S.J. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single Residue on the WPD-Loop Affects the pH Dependency of Catalysis in Protein Tyrosine Phosphatases.
Jacs Au, 1, 2021
|
|
5W8F
 
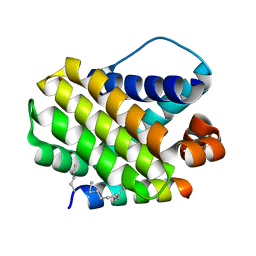 | | Crystal structure of human Mcl-1 in complex with modified Bim BH3 peptide SAH-MS1-14 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, modified Bim BH3 peptide SAH-MS1-14 | | Authors: | Rezaei Araghi, R, Jenson, J.M, Grant, R.A, Keating, A.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Iterative optimization yields Mcl-1-targeting stapled peptides with selective cytotoxicity to Mcl-1-dependent cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4GJ1
 
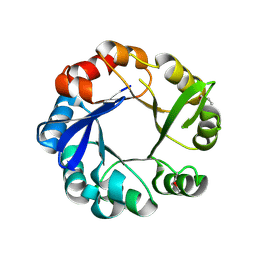 | | Crystal structure of 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase (hisA). | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase | | Authors: | Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-09 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Crystal structure of 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase (hisA).
To be Published
|
|
5WRM
 
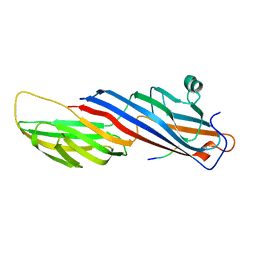 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y658 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
3AK3
 
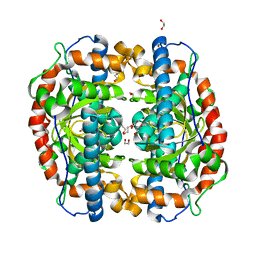 | | Superoxide dismutase from Aeropyrum pernix K1, Fe-bound form | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Superoxide dismutase [Mn/Fe] | | Authors: | Nakamura, T, Uegaki, K. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of the cambialistic superoxide dismutase from Aeropyrum pernix K1 - insights into the enzyme mechanism and stability
Febs J., 278, 2011
|
|
6MBO
 
 | | GLP Methyltransferase with Inhibitor EML741-P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
6MC1
 
 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | Authors: | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | Deposit date: | 2018-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
1MXR
 
 | | High resolution structure of Ribonucleotide reductase R2 from E. coli in its oxidised (Met) form | | Descriptor: | FE (III) ION, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Andersson, M.A, Hogbom, M, Nordlund, P. | | Deposit date: | 2002-10-03 | | Release date: | 2003-03-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Displacement of the tyrosyl radical cofactor in ribonucleotide reductase obtained by single-crystal high-field EPR and 1.4-A x-ray data.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
7M13
 
 | | Crystal structure of CJ1428, a GDP-D-GLYCERO-L-GLUCO-HEPTOSE SYNTHASE from campylobacter jejuni in the presence of NADPH | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, MAGNESIUM ION, ... | | Authors: | Anderson, T.K, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
3O7M
 
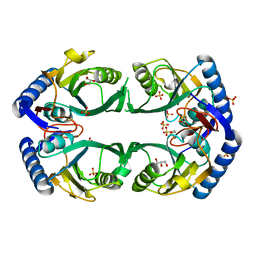 | | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor'
TO BE PUBLISHED
|
|
5XD0
 
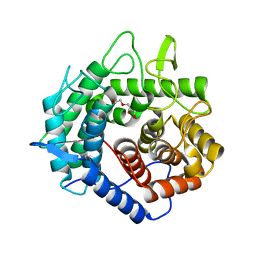 | | Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glucanase, TRIETHYLENE GLYCOL | | Authors: | Baek, S.C, Ho, T.-H, Kang, L.-W, Kim, H. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Improvement of enzyme activity of beta-1,3-1,4-glucanase from Paenibacillus sp. X4 by error-prone PCR and structural insights of mutated residues.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
1U17
 
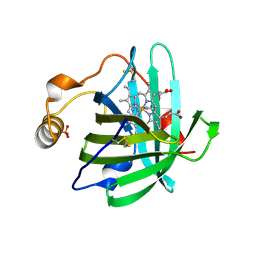 | |
1TPH
 
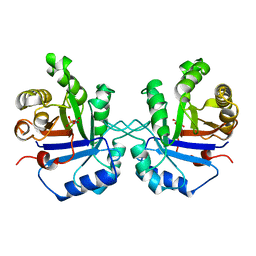 | | 1.8 ANGSTROMS CRYSTAL STRUCTURE OF WILD TYPE CHICKEN TRIOSEPHOSPHATE ISOMERASE-PHOSPHOGLYCOLOHYDROXAMATE COMPLEX | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-22 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of recombinant chicken triosephosphate isomerase-phosphoglycolohydroxamate complex at 1.8-A resolution.
Biochemistry, 33, 1994
|
|
5XCZ
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium in complex with cellobiose at 2.1 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4G31
 
 | | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
5TVM
 
 | | Crystal structure of Trypanosoma brucei AdoMetDC/prozyme heterodimer | | Descriptor: | 1,4-DIAMINOBUTANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Volkov, O.A, Chen, Z, Tomchick, D.R, Phillips, M.A. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
6QC5
 
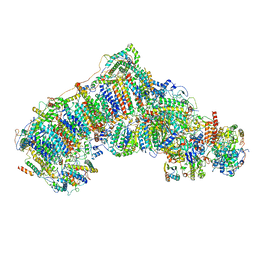 | | Ovine respiratory complex I FRC closed class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-26 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
3LDV
 
 | | 1.77 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Glass, E.M, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | 1.77 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
5TZH
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 2A IN COMPLEX WITH 3,3-difluoro-1-[(4-fluorophenyl)carbonyl]-5-{5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl}piperidine | | Descriptor: | MAGNESIUM ION, ZINC ION, [(5S)-3,3-difluoro-5-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl](4-fluorophenyl)methanone, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
5TZW
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 2A IN COMPLEX WITH 1-[(3,4-difluorophenyl)carbonyl]-3,3-difluoro-5-{5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl}piperidine | | Descriptor: | MAGNESIUM ION, ZINC ION, [(5S)-3,3-difluoro-5-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl](3,4-difluorophenyl)methanone, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
