6PIV
 
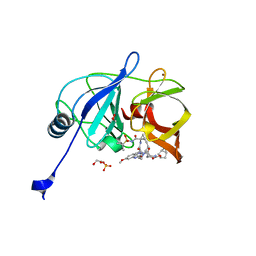 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-7 (NR03-77) | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PJ0
 
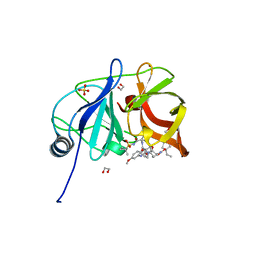 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-5 (NR01-97) | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4 Aprotease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6UBU
 
 | |
6U2D
 
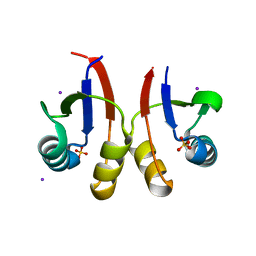 | | PmtCD peptide exporter basket domain | | Descriptor: | ABC transporter ATP-binding protein, IODIDE ION, SULFATE ION | | Authors: | Zeytuni, N, Strynadka, N.C.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
6PGZ
 
 | | MTSL labelled T4 lysozyme pseudo-wild type V75C mutant | | Descriptor: | CHLORIDE ION, Endolysin, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
6P9U
 
 | | Crystal structure of human thrombin mutant W215A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, ZINC ION | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Di Cera, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
6PEJ
 
 | | Structure of sorbitol dehydrogenase from Sinorhizobium meliloti 1021 bound to sorbitol | | Descriptor: | Sorbitol dehydrogenase (L-iditol 2-dehydrogenase), sorbitol | | Authors: | Bailey-Elkin, B.A, Kohlmeier, M.G, Oresnik, I.J, Mark, B.L. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the sorbitol dehydrogenase SmoS from Sinorhizobium meliloti 1021
Acta Crystallogr.,Sect.D, 77, 2021
|
|
6PJB
 
 | | HIV-1 Protease NL4-3 WT in Complex with Lopinavir | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PJN
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-41 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3S,5S)-3-hydroxy-5-({1-[(methoxycarbonyl)amino]cyclopentane-1-carbonyl}amino)-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6U0C
 
 | |
6PLZ
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, closed state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PHV
 
 | | SpAga galactose product complex structure | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-galactosidase, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
6UFL
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Cordeiro, R.L, Domingues, M.N, Vieira, P.S, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
7K4M
 
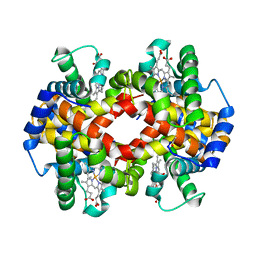 | | Crystal structure of MetAP2 Modified Hemoglobin S | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Musayev, F.N, Safo, M.K, Light, D.R. | | Deposit date: | 2020-09-15 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MetAP2 inhibition modifies hemoglobin S to delay polymerization and improves blood flow in sickle cell disease.
Blood Adv, 5, 2021
|
|
6PIW
 
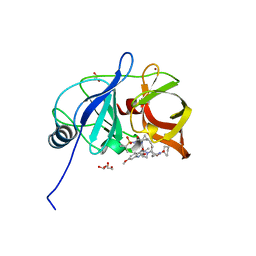 | |
6PJ1
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-4(AJ-74) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PLY
 
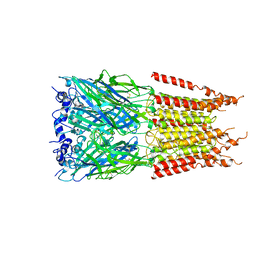 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, open state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6UKE
 
 | |
6U52
 
 | | Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB) Complexed with Sudan ebolavirus Nucleoprotein C-terminal Domain 634-738 | | Descriptor: | Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB), CHLORIDE ION, Nucleoprotein | | Authors: | Taylor, A.B, Sherwood, L.J, Hart, P.J, Hayhurst, A. | | Deposit date: | 2019-08-26 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Paratope Duality and Gullying are Among the Atypical Recognition Mechanisms Used by a Trio of Nanobodies to Differentiate Ebolavirus Nucleoproteins.
J.Mol.Biol., 431, 2019
|
|
6OZ1
 
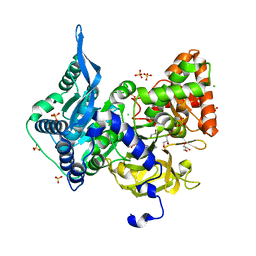 | | Crystal structure of the adenylation (A) domain of the carboxylate reductase (CAR) GR01_22995 from Mycobacterium chelonae | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Fedorchuk, T, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | One-Pot Biocatalytic Transformation of Adipic Acid to 6-Aminocaproic Acid and 1,6-Hexamethylenediamine Using Carboxylic Acid Reductases and Transaminases.
J.Am.Chem.Soc., 142, 2020
|
|
6UFZ
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) | | Descriptor: | Glyco_hydro_cc domain-containing protein | | Authors: | Cordeiro, R.L, Domingues, M.N, Vieira, P.S, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UBH
 
 | | Structure of the MM7 Erbin PDZ variant in complex with a high-affinity peptide | | Descriptor: | Erbin, SODIUM ION, peptide | | Authors: | Singer, A.U, Teyra, J, McLaughlin, M, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-09-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comprehensive Assessment of the Relationship Between Site -2 Specificity and Helix alpha 2 in the Erbin PDZ Domain.
J.Mol.Biol., 433, 2021
|
|
6UC7
 
 | | Structure of guanine riboswitch bound to N2-acetyl guanine | | Descriptor: | COBALT HEXAMMINE(III), N-(6-oxo-6,9-dihydro-3H-purin-2-yl)acetamide, guanine riboswitch | | Authors: | Matyjasik, M.M, Batey, R.T. | | Deposit date: | 2019-09-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | High Affinity Binding of N2-Modified Guanine Derivatives Significantly Disrupts the Ligand Binding Pocket of the Guanine Riboswitch.
Molecules, 25, 2020
|
|
6P5C
 
 | | Bacillus Fragment DNA polymerase mutant I716M | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*TP*GP*AP*TP*CP*G)-3'), DNA polymerase I, ... | | Authors: | Wu, E.Y. | | Deposit date: | 2019-05-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The importance of Ile716 toward the mutagenicity of 8-Oxo-2'-deoxyguanosine with Bacillus fragment DNA polymerase.
DNA Repair (Amst.), 89, 2020
|
|
6UN8
 
 | | Wild type ANT bound to neomycin | | Descriptor: | Aminoglycoside NucleotidylTransferase, MAGNESIUM ION, NEOMYCIN | | Authors: | Cuneo, M.J, Selvaraj, B. | | Deposit date: | 2019-10-11 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | "Catch and Release": a Variation of the Archetypal NucleotidylTransfer Reaction
ACS Catal, 10, 2020
|
|
