8RXX
 
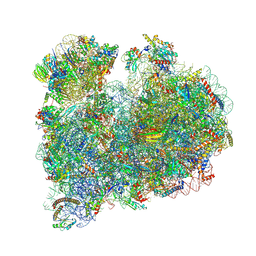 | |
8RSA
 
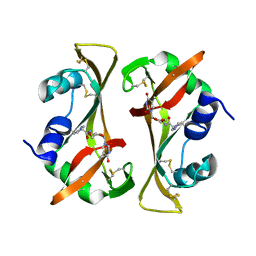 | |
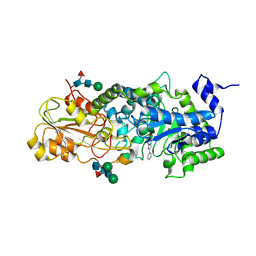
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
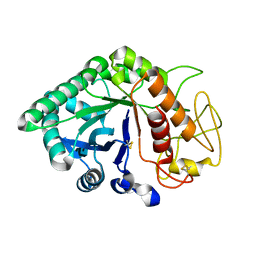
1LQ2
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional Structure of the Barley {beta}-D-Glucan Glucohydrolase in Complex with a Transition State Mimic.
J.Biol.Chem., 279, 2004
|
|
1LJY
 
 | | Crystal Structure of a Novel Regulatory 40 kDa Mammary Gland Protein (MGP-40) secreted during Involution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MGP-40 | | Authors: | Mohanty, A.K, Singh, G, Paramasivam, M, Saravanan, K, Jabeen, T, Sharma, S, Yadav, S, Kaur, P, Kumar, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-04-23 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Novel Regulatory 40 kDa Mammary Gland Protein (MGP-40) secreted during Involution
J.Biol.Chem., 278, 2003
|
|
8HV7
 
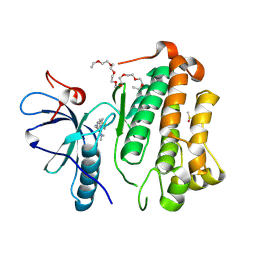 | | Crystal structure of EGFR_TMX in complex with covalently bound fragment 9 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DIMETHYL SULFOXIDE, Epidermal growth factor receptor, ... | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
8HV8
 
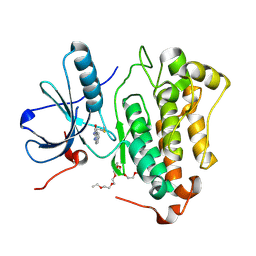 | | Crystal structure of EGFR_TMX in complex with covalently bound fragment 10 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Epidermal growth factor receptor, ~{N}-pyrazolo[1,5-a]pyridin-2-ylprop-2-enamide | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
2I3I
 
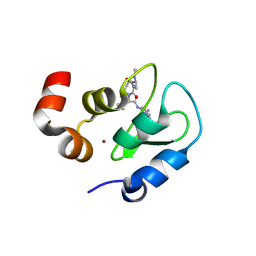 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | (3R,6R,9AR)-2,2-DIMETHYL-6-[(N-METHYL-L-ALANYL)AMINO]-N-(3-METHYL-1-PHENYL-1H-PYRAZOL-5-YL)-5-OXO-2,3,5,6,9,9A-HEXAHYDRO[1,3]THIAZOLO[3,2-A]AZEPINE-3-CARBOXAMIDE, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fairbrother, W.J, Franklin, M.C. | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs.
Acs Chem.Biol., 1, 2006
|
|
5JOO
 
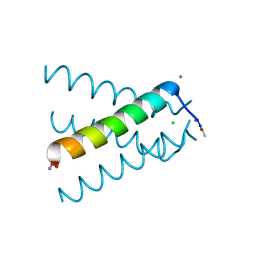 | | XFEL structure of influenza A M2 wild type TM domain at low pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2016-05-02 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2IIT
 
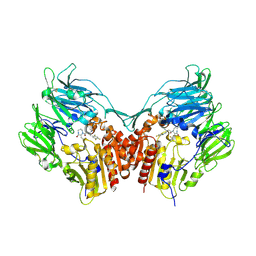 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | Descriptor: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-(2,2,2-TRIFLUOROETHYL)-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T, Weber, A.E. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2J1E
 
 | |
4W2G
 
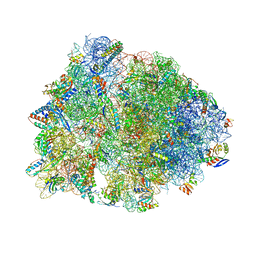 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (soaked), mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
5K9J
 
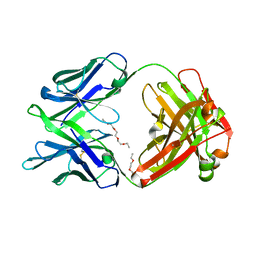 | | Crystal structure of multidonor HV6-1-class broadly neutralizing Influenza A antibody 56.a.09 isolated following H5 immunization. | | Descriptor: | 56.a.09 heavy chain, 56.a.09 light chain, POLYETHYLENE GLYCOL (N=34) | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-31 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
8BK0
 
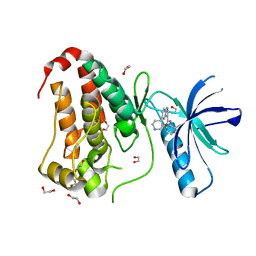 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904 | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chlorophenyl)-6-piperidin-4-yl-imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Zhubi, R, Gerninghaus, J, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904
To Be Published
|
|
5DA7
 
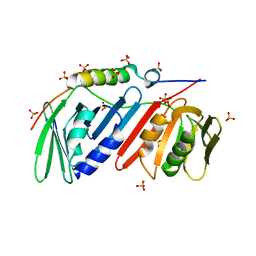 | | monomeric PCNA bound to a small protein inhibitor | | Descriptor: | DNA polymerase sliding clamp 1, Proliferating cell nuclear antigen, SULFATE ION, ... | | Authors: | Ladner, J.E, Altieri, A.S, Kelman, Z. | | Deposit date: | 2015-08-19 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A small protein inhibits proliferating cell nuclear antigen by breaking the DNA clamp.
Nucleic Acids Res., 44, 2016
|
|
2VN6
 
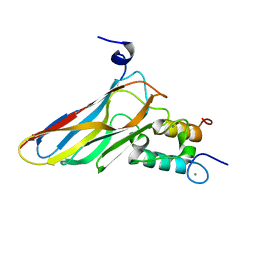 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
2I8E
 
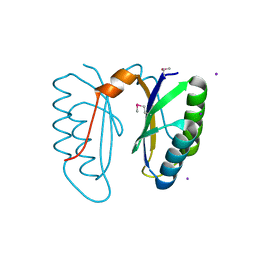 | | Structure of SSO1404, a predicted DNA repair-associated protein from Sulfolobus solfataricus P2 | | Descriptor: | Hypothetical protein, IODIDE ION | | Authors: | Wang, S, Zimmerman, M.D, Kudritska, M, Chruszcz, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats.
J.Biol.Chem., 283, 2008
|
|
6T2C
 
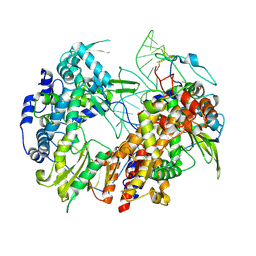 | | Bat Influenza A polymerase recycling complex | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2019-10-08 | | Release date: | 2020-04-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
2VEO
 
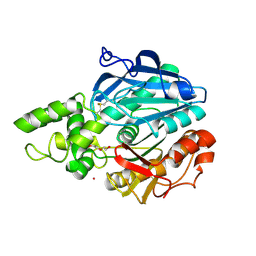 | | X-ray structure of Candida antarctica lipase A in its closed state. | | Descriptor: | GLYCEROL, LIPASE A, TETRAETHYLENE GLYCOL, ... | | Authors: | Ericsson, D.J, Kasrayan, A, Johansson, P, Bergfors, T, Sandstrom, A.G, Backvall, J.E, Mowbray, S.L. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure of Candida Antarctica Lipase a Shows a Novel Lid Structure and a Likely Mode of Interfacial Activation.
J.Mol.Biol., 376, 2008
|
|
6ERJ
 
 | | Self-complemented FimA subunit from Salmonella enterica | | Descriptor: | ACETIC ACID, Type-1 fimbrial protein, a chain | | Authors: | Zyla, D.S, Prota, A, Capitani, G, Glockshuber, R. | | Deposit date: | 2017-10-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 294, 2019
|
|
1U67
 
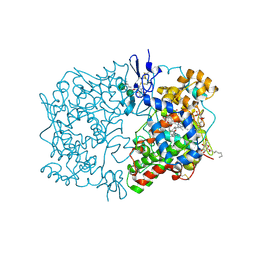 | | Crystal Structure of Arachidonic Acid Bound to a Mutant of Prostagladin H Synthase-1 that Forms Predominantly 11-HPETE. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Harman, C.A, Rieke, C.J, Garavito, R.M, Smith, W.L. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of arachidonic Acid bound to a mutant of prostaglandin endoperoxide h synthase-1 that forms predominantly 11-hydroperoxyeicosatetraenoic Acid.
J.Biol.Chem., 279, 2004
|
|
5XPN
 
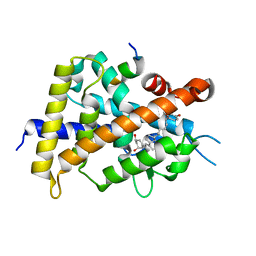 | | Crystal structure of VDR-LBD complexed with 25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{S})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XDQ
 
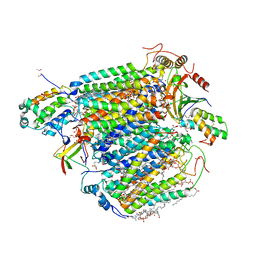 | | Bovine heart cytochrome c oxidase in the fully oxidized state with pH 7.3 at 1.77 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-03-29 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of bovine cytochrome c oxidase crystallized at a neutral pH using a fluorinated detergent.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4V62
 
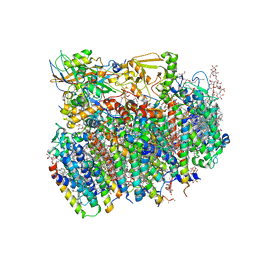 | | Crystal Structure of cyanobacterial Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Guskov, A, Gabdulkhakov, A, Kern, J, Broser, M, Zouni, A, Saenger, W. | | Deposit date: | 2008-01-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride
Nat.Struct.Mol.Biol., 16, 2009
|
|
2IDN
 
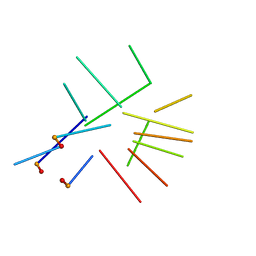 | | NMR structure of a new modified Thrombin Binding Aptamer containing a 5'-5' inversion of polarity site | | Descriptor: | 3'-D(P*GP*G*T)-5'-5'-D(P*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3' | | Authors: | Randazzo, A, Martino, L, Virno, A, Mayol, L, Giancola, C. | | Deposit date: | 2006-09-15 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A new modified thrombin binding aptamer containing a 5'-5' inversion of polarity site.
Nucleic Acids Res., 34, 2006
|
|
