2I87
 
 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | D-alanine-D-alanine ligase, SULFATE ION | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DVE
 
 | |
4QBJ
 
 | | Crystal structure of N-myristoyl transferase from Aspergillus fumigatus complexed with a synthetic inhibitor | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, Glycylpeptide N-tetradecanoyltransferase, S-(2-OXO)PENTADECYLCOA, ... | | Authors: | Suzuki, M, Shimada, T. | | Deposit date: | 2014-05-08 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of N-myristoyltransferase from Aspergillus fumigatus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2KXM
 
 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostmycin complex | | Descriptor: | RIBOSTAMYCIN, RNA (27-MER) | | Authors: | Duchardt-Ferner, E, Weigand, J.E, Ohlenschlager, O, Schmidtke, S.R, Suess, B, Wohnert, J. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2DTO
 
 | |
4B0T
 
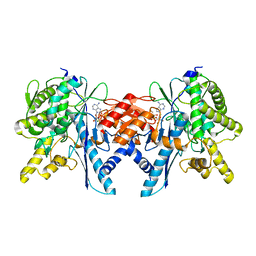 | | Structure of the Pup Ligase PafA of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
2DXT
 
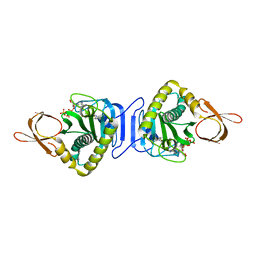 | |
7VHV
 
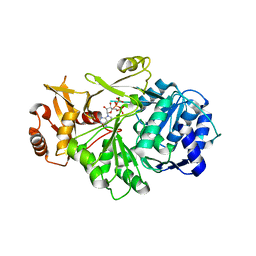 | | Crystal structure of S. aureus D-alanine alanyl carrier protein ligase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-alanine--D-alanyl carrier protein ligase, MAGNESIUM ION | | Authors: | Lee, B.J, Lee, I.-G, Im, H.G, Yoon, H.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional analysis of the D-alanyl carrier protein ligase DltA from Staphylococcus aureus Mu50.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8Z7D
 
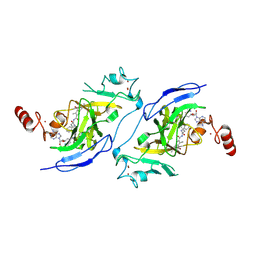 | | Structure of G9a in complex with compound 9a | | Descriptor: | 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-[(phenylmethyl)amino]hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
8Z7E
 
 | | Structure of G9a in complex with compound 9b | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ZINC ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
8Z7C
 
 | | Structure of G9a in complex with compound 7i | | Descriptor: | 3,6,6-trimethyl-~{N}-[(2~{S})-1-[[4-(1-methylpiperidin-4-yl)oxyphenyl]amino]-1-oxidanylidene-hexan-2-yl]-4-oxidanylidene-5,7-dihydro-1~{H}-indole-2-carboxamide, Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
1X9N
 
 | | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA | | Descriptor: | 5'-phosphorylated DNA, ADENOSINE MONOPHOSPHATE, DNA ligase I, ... | | Authors: | Pascal, J.M, O'Brien, P.J, Tomkinson, A.E, Ellenberger, T. | | Deposit date: | 2004-08-23 | | Release date: | 2004-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA ligase I completely encircles and partially unwinds nicked DNA.
Nature, 432, 2004
|
|
5W97
 
 | | Crystal Structure of CO-bound Cytochrome c Oxidase determined by Serial Femtosecond X-Ray Crystallography at Room Temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I, Zatsepin, N.A, Grant, T.D, Fromme, P, Fromme, R. | | Deposit date: | 2017-06-22 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8TQM
 
 | |
5WAU
 
 | | Crystal Structure of CO-bound Cytochrome c Oxidase determined by Synchrotron X-Ray Crystallography at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Fromme, R, Ishigami, I, Yeh, S.Y, Zatsepin, N, Grant, T, Fromme, P, Rousseau, D. | | Deposit date: | 2017-06-27 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3VMM
 
 | |
2OOA
 
 | |
2E65
 
 | |
2I8C
 
 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase, MAGNESIUM ION, ... | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
9GIM
 
 | |
9GIK
 
 | |
3A7A
 
 | | Crystal structure of E. coli lipoate-protein ligase A in complex with octyl-amp and apoH-protein | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine cleavage system H protein, Lipoate-protein ligase A, ... | | Authors: | Fujiwara, K, Hosaka, H, Nakagawa, A. | | Deposit date: | 2009-09-20 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A.
J.Biol.Chem., 285, 2010
|
|
7XUB
 
 | | Structure of G9a in complex with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUC
 
 | | Structure of G9a in complex with compound 11a | | Descriptor: | 1,2-ETHANEDIOL, 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-phenylazanyl-hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
2GWD
 
 | | Crystal structure of plant glutamate cysteine ligase in complex with Mg2+ and L-glutamate | | Descriptor: | ACETATE ION, GLUTAMIC ACID, Glutamate cysteine ligase, ... | | Authors: | Hothorn, M, Wachter, A, Gromes, R, Stuwe, T, Rausch, T, Scheffzek, K. | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for the redox control of plant glutamate cysteine ligase.
J.Biol.Chem., 281, 2006
|
|
