6JWR
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS wild type with Pteroate | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6LJJ
 
 | | Swine dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial isoform 1, ... | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6ONK
 
 | | Dehaloperoxidase B in complex with substrate 4-Cl-cresol | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-methylphenol, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The multifunctional globin dehaloperoxidase strikes again: Simultaneous peroxidase and peroxygenase mechanisms in the oxidation of EPA pollutants.
Arch.Biochem.Biophys., 673, 2019
|
|
6ONR
 
 | | Dehaloperoxidase B in complex with substrate 4-methyl-cresol | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dimethylphenol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The multifunctional globin dehaloperoxidase strikes again: Simultaneous peroxidase and peroxygenase mechanisms in the oxidation of EPA pollutants.
Arch.Biochem.Biophys., 673, 2019
|
|
6OO1
 
 | | Dehaloperoxidase B in complex with substrate o-cresol | | Descriptor: | 1,2-ETHANEDIOL, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2019-04-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | The multifunctional globin dehaloperoxidase strikes again: Simultaneous peroxidase and peroxygenase mechanisms in the oxidation of EPA pollutants.
Arch.Biochem.Biophys., 673, 2019
|
|
6OO6
 
 | | Dehaloperoxidase B in complex with substrate p-cresol | | Descriptor: | 1,2-ETHANEDIOL, Dehaloperoxidase B, P-CRESOL, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multifunctional globin dehaloperoxidase strikes again: Simultaneous peroxidase and peroxygenase mechanisms in the oxidation of EPA pollutants.
Arch.Biochem.Biophys., 673, 2019
|
|
6RWV
 
 | | Structure of apo-LmCpfC | | Descriptor: | Ferrochelatase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6386379 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
6L0G
 
 | | Crystal structure of dihydroorotase in complex with malate at pH6 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
5N0E
 
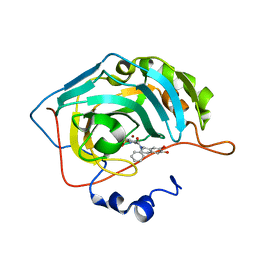 | | Crystal structure of human carbonic anhydrase II in complex with (S)-4-(6,7-dihydroxy-1-phenyl-3,4-tetrahydroisoquinoline-1H-2-carbonyl)benzenesulfonamide. | | Descriptor: | 4-[[(1~{S})-6,7-bis(oxidanyl)-1-phenyl-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing Molecular Interactions between Human Carbonic Anhydrases (hCAs) and a Novel Class of Benzenesulfonamides.
J. Med. Chem., 60, 2017
|
|
6ONG
 
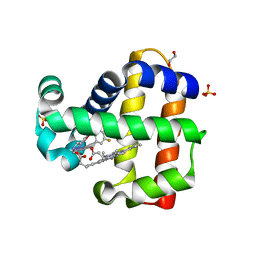 | | Dehaloperoxidate B in complex with substrate 4-F-cresol | | Descriptor: | 1,2-ETHANEDIOL, 4-fluoro-2-methylphenol, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T, McGuire, A. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The multifunctional globin dehaloperoxidase strikes again: Simultaneous peroxidase and peroxygenase mechanisms in the oxidation of EPA pollutants.
Arch.Biochem.Biophys., 673, 2019
|
|
6ONX
 
 | | Dehaloperoxidase B in complex with substrate 4-Br-cresol | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-2-methylphenol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The multifunctional globin dehaloperoxidase strikes again: Simultaneous peroxidase and peroxygenase mechanisms in the oxidation of EPA pollutants.
Arch.Biochem.Biophys., 673, 2019
|
|
6O50
 
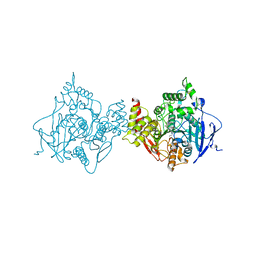 | | Binary complex of native hAChE with BW284c51 | | Descriptor: | 4-(5-{4-[DIMETHYL(PROP-2-ENYL)AMMONIO]PHENYL}-3-OXOPENTYL)-N,N-DIMETHYL-N-PROP-2-ENYLBENZENAMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
6KCM
 
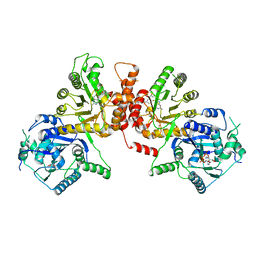 | | Crystal structure of Plasmodium falciparum HPPK-DHPS S436F/A437G/A613S with pterin and p-hydrobenzoate | | Descriptor: | 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6RFT
 
 | | Crystal structure of Eis2 from Mycobacterium abscessus bound to Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Uncharacterized N-acetyltransferase D2E36_21790 | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6RGU
 
 | | Photorhabdus asymbiotica lectin PHL in complex with 3-O-methyl-D-glucose | | Descriptor: | 3-O-methyl-beta-D-glucopyranose, CHLORIDE ION, Lectin PHL, ... | | Authors: | Houser, J, Fujdiarova, E, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2019-04-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
6RFX
 
 | | Crystal structure of Eis2 from Mycobacterium abscessus | | Descriptor: | ACETATE ION, CITRIC ACID, Eis2, ... | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6LPK
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 293 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
9E9B
 
 | | Crystal structure of L. monocytogenes MenD with Mg2+ and ThDP bound | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Klein, M, Given, F.M, Ho, N.A.T, Allison, T.M, Johnston, J.M. | | Deposit date: | 2024-11-08 | | Release date: | 2025-07-30 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structures of Listeria monocytogenes MenD in ThDP-bound and in-crystallo captured intermediate I-bound forms.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
6LT8
 
 | | HSP90 in complex with KW-2478 | | Descriptor: | 2-[2-ethyl-6-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]carbonyl-3,5-bis(oxidanyl)phenyl]-~{N},~{N}-bis(2-methoxyethyl)ethanamide, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Anti-NSCLC activity in vitro of Hsp90 N inhibitor KW-2478 and complex crystal structure determination of Hsp90 N -KW-2478.
J.Struct.Biol., 213, 2021
|
|
2YER
 
 | | Synthesis and evaluation of triazolones as checkpoint kinase 1 inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 5-(HYDROXYMETHYL)-8-(1H-PYRROL-2-YL)-2H-[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Vallentine, A, White, A, Otterbein, L. | | Deposit date: | 2011-03-30 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis and Evaluation of Triazolones as Checkpoint Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2YEX
 
 | | Synthesis and evaluation of triazolones as checkpoint kinase 1 inhibitors | | Descriptor: | 5-METHYL-8-(1H-PYRROL-2-YL)[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1(2H)-ONE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Vallentine, A, White, A. | | Deposit date: | 2011-03-31 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis and Evaluation of Triazolones as Checkpoint Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6MAB
 
 | | 1.7A resolution structure of RsbU from Chlamydia trachomatis (periplasmic domain) | | Descriptor: | ISOPROPYL ALCOHOL, Sigma regulatory family protein-PP2C phosphatase | | Authors: | Dmitriev, A, Lovell, S, Battaile, K.P, Soules, K, Hefty, P.S. | | Deposit date: | 2018-08-27 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and ligand binding analyses of the periplasmic sensor domain of RsbU in Chlamydia trachomatis support a role in TCA cycle regulation.
Mol.Microbiol., 113, 2020
|
|
6MBB
 
 | | Human Bfl-1 in complex with the designed peptide dF1 | | Descriptor: | Bcl-2-related protein A1, dF1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6LR9
 
 | | HSP90 in complex with Debio0932 | | Descriptor: | 2-[[6-(dimethylamino)-1,3-benzodioxol-5-yl]sulfanyl]-1-[2-(2,2-dimethylpropylamino)ethyl]imidazo[4,5-c]pyridin-4-amine, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Complex crystal structure determination and anti-non-small-cell lung cancer activity of the Hsp90 N inhibitor Debio0932.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6LTK
 
 | | HSP90 in complex with SNX-2112 | | Descriptor: | 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Complex Crystal Structure Determination and in vitro Anti-non-small Cell Lung Cancer Activity of Hsp90 N Inhibitor SNX-2112.
Front Cell Dev Biol, 9, 2021
|
|
