8HEW
 
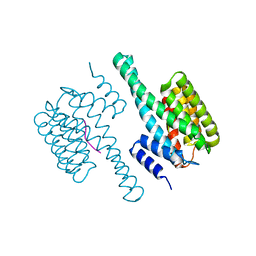 | | Potato 14-3-3 St14f | | Descriptor: | 14-3-3 protein, StFDL1 peptide | | Authors: | Taoka, K, Kawahara, I, Shinya, S, Harada, K, Muranaka, T, Furuita, K, Nakagawa, A, Fujiwara, T, Tsuji, H, Kojima, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multifunctional chemical inhibitors of the florigen activation complex discovered by structure-based high-throughput screening.
Plant J., 112, 2022
|
|
4HI2
 
 | |
4HI1
 
 | |
4I2N
 
 | |
4I46
 
 | |
4H60
 
 | |
4H18
 
 | | Three dimensional structure of corynomycoloyl tranferase C | | Descriptor: | Cmt1, MAGNESIUM ION | | Authors: | Huc, E, de Sousa D'Auria, C, Li de la Sierra-Gallay, I, Salmeron, C.H, van Tilbeurgh, H, Bayan, N, Houssin, C.H, Daffe, M, Tropis, M. | | Deposit date: | 2012-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Identification of a mycoloyl transferase selectively involved in o-acylation of polypeptides in corynebacteriales.
J.Bacteriol., 195, 2013
|
|
5YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH HEXAFLUOROACETONE | | Descriptor: | 1,1,1,3,3,3-HEXAFLUOROPROPANEDIOL, PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
6SWZ
 
 | |
6SX4
 
 | |
2NUW
 
 | |
6TEP
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
2PH9
 
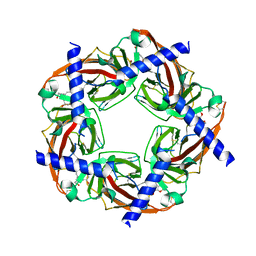 | | Galanthamine bound to an ACh-binding Protein | | Descriptor: | (-)-GALANTHAMINE, Soluble acetylcholine receptor, TETRAETHYLENE GLYCOL | | Authors: | Hansen, S.B, Taylor, P. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Galanthamine and non-competitive inhibitor binding to ACh-binding protein: evidence for a binding site on non-alpha-subunit interfaces of heteromeric neuronal nicotinic receptors.
J.Mol.Biol., 369, 2007
|
|
2PGZ
 
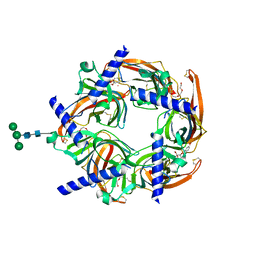 | | Crystal structure of Cocaine bound to an ACh-Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COCAINE, Soluble acetylcholine receptor, ... | | Authors: | Hansen, S.B, Taylor, P. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Galanthamine and non-competitive inhibitor binding to ACh-binding protein: evidence for a binding site on non-alpha-subunit interfaces of heteromeric neuronal nicotinic receptors.
J.Mol.Biol., 369, 2007
|
|
2PVS
 
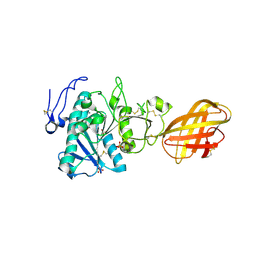 | | Structure of human pancreatic lipase related protein 2 mutant N336Q | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 2, SULFATE ION | | Authors: | Spinelli, S, Eydoux, C, Carriere, F, Cambillau, C. | | Deposit date: | 2007-05-10 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
6LY8
 
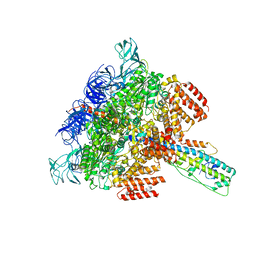 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
2NUY
 
 | |
6TEQ
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-galactopyranose, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
2NUX
 
 | |
6TER
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with Galactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
4G7X
 
 | |
4G4O
 
 | | MutM containing M77A mutation bound to oxoG-containing DNA | | Descriptor: | DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*AP*GP*(TX2)P*CP*TP*AP*CP*C)-3'), DNA (5'-D(P*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Analysis of DNA Helix Invasion by the Bacterial 8-Oxoguanine DNA Glycosylase MutM.
J.Biol.Chem., 288, 2013
|
|
4G4R
 
 | | MutM containing F114A mutation bound to oxoG-containing DNA | | Descriptor: | DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*AP*GP*(TX2)P*CP*TP*AP*CP*C)-3'), DNA (5'-D(P*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Analysis of DNA Helix Invasion by the Bacterial 8-Oxoguanine DNA Glycosylase MutM.
J.Biol.Chem., 288, 2013
|
|
2PO3
 
 | | Crystal Structure Analysis of DesI in the presence of its TDP-sugar product | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 4-dehydrase | | Authors: | Burgie, E.S, Holden, H.M. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Architecture of DesI: A Key Enzyme in the Biosynthesis of Desosamine
Biochemistry, 46, 2007
|
|
4G4Q
 
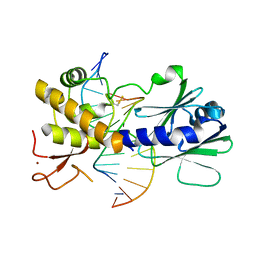 | | MutM containing F114A mutation bound to undamaged DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*AP*GP*(TX2)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Biochemical Analysis of DNA Helix Invasion by the Bacterial 8-Oxoguanine DNA Glycosylase MutM.
J.Biol.Chem., 288, 2013
|
|
