8EGW
 
 | | Complex of Fat4(EC1-4) bound to Dchs1(EC1-3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Medina, E, Luca, V.C. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the planar cell polarity cadherins Fat4 and Dachsous1.
Nat Commun, 14, 2023
|
|
1NHI
 
 | | Crystal structure of N-terminal 40KD MutL (LN40) complex with ADPnP and one potassium | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein mutL, MAGNESIUM ION, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
3UL7
 
 | | Crystal structure of the TV3 mutant F63W | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
2XXR
 
 | | Crystal structure of the GluK2 (GluR6) wild-type LBD dimer in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-11 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
2Y0M
 
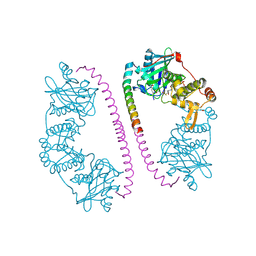 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN DOSAGE COMPENSATION FACTORS MSL1 AND MOF | | Descriptor: | ACETYL COENZYME *A, MALE-SPECIFIC LETHAL 1 HOMOLOG, PROBABLE HISTONE ACETYLTRANSFERASE MYST1, ... | | Authors: | Kadlec, J, Hallacli, E, Lipp, M, Holz, H, Sanchez Weatherby, J, Cusack, S, Akhtar, A. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Mof and Msl3 Recruitment Into the Dosage Compensation Complex by Msl1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XQV
 
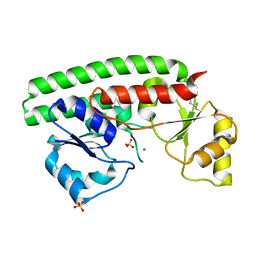 | | The X-ray structure of the Zn(II) bound ZnuA from Salmonella enterica | | Descriptor: | SULFATE ION, ZINC ABC TRANSPORTER, PERIPLASMIC ZINC-BINDING PROTEIN, ... | | Authors: | Alaleona, F, Ilari, A, Chiancone, E, Battistoni, A, Petrarca, P. | | Deposit date: | 2010-09-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The X-Ray Structure of the Zinc Transporter Znua from Salmonella Enterica Discloses a Unique Triad of Zinc Coordinating Histidines.
J.Mol.Biol., 409, 2011
|
|
2XXV
 
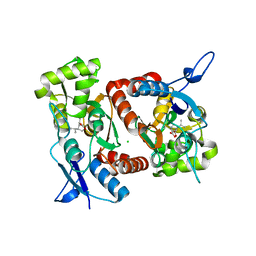 | | Crystal structure of the GluK2 (GluR6) M770K LBD dimer in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
2Y0N
 
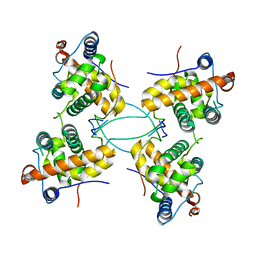 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN DOSAGE COMPENSATION FACTORS MSL1 AND MSL3 | | Descriptor: | MALE-SPECIFIC LETHAL 1 HOMOLOG, MALE-SPECIFIC LETHAL 3 HOMOLOG | | Authors: | Kadlec, J, Hallacli, E, Lipp, M, Holz, H, Sanchez Weatherby, J, Cusack, S, Akhtar, A. | | Deposit date: | 2010-12-04 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Mof and Msl3 Recruitment Into the Dosage Compensation Complex by Msl1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
5XTO
 
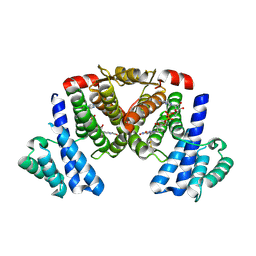 | |
2XLC
 
 | | Acetyl xylan esterase from Bacillus pumilus CECT5072 bound to paraoxon | | Descriptor: | ACETYL XYLAN ESTERASE, DIETHYL PHOSPHONATE | | Authors: | Gil-Ortiz, F, Montoro-Garcia, S, Polo, L.M, Rubio, V, Sanchez-Ferrer, A. | | Deposit date: | 2010-07-20 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Cephalosporin Deacetylating Enzyme Acetyl Xylan Esterase Bound to Paraoxon Explains the Low Sensitivity of This Serine Hydrolase to Organophosphate Inactivation.
Biochem.J., 436, 2011
|
|
3UCJ
 
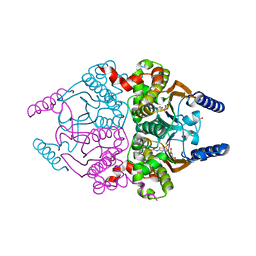 | | Coccomyxa beta-carbonic anhydrase in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CHLORIDE ION, Carbonic anhydrase, ... | | Authors: | Huang, S, Hainzl, T, Sauer-Eriksson, A.E. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of [beta]-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide.
Plos One, 6, 2011
|
|
2XQW
 
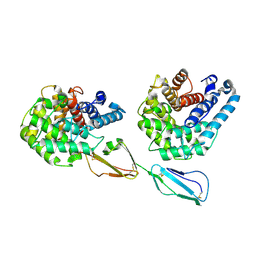 | | Structure of Factor H domains 19-20 in complex with complement C3d | | Descriptor: | COMPLEMENT C3, COMPLEMENT FACTOR H | | Authors: | Kajander, T, Lehtinen, M.J, Hyvarinen, S, Bhattacharjee, A, Leung, E, Isenman, D.E, Meri, S, Jokiranta, T.S, Goldman, A. | | Deposit date: | 2010-09-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Dual Interaction of Factor H with C3D and Glycosaminoglycans in Host-Nonhost Discrimination by Complement.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4K7J
 
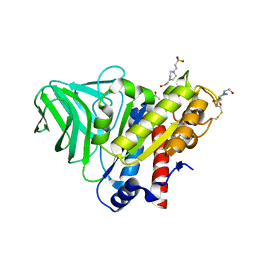 | | Peptidoglycan O-acetylesterase in action, 5 min | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GDSL-like Lipase/Acylhydrolase family protein | | Authors: | Williams, A.H, Gomperts Boneca, I. | | Deposit date: | 2013-04-17 | | Release date: | 2014-09-03 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Visualization of a substrate-induced productive conformation of the catalytic triad of the Neisseria meningitidis peptidoglycan O-acetylesterase reveals mechanistic conservation in SGNH esterase family members.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8R8K
 
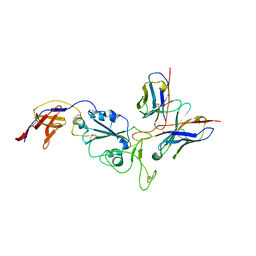 | |
2XSQ
 
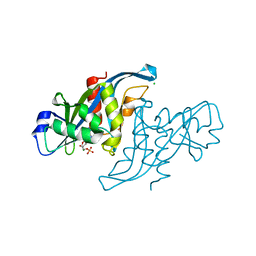 | | Crystal structure of human Nudix motif 16 (NUDT16) in complex with IMP and magnesium | | Descriptor: | CHLORIDE ION, INOSINIC ACID, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van den Berg, S, Wahlberg, E, Weigelt, J, Nordlund, P. | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for the Specificity of Human Nudt16 and its Regulation by Inosine Monophosphate.
Plos One, 10, 2015
|
|
2XXW
 
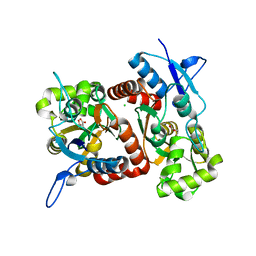 | | Crystal structure of the GluK2 (GluR6) D776K LBD dimer in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
4K3U
 
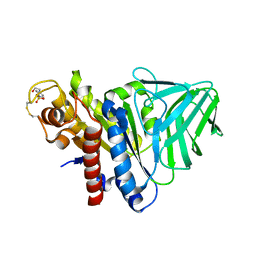 | | Peptidoglycan O-acetylesterase in action, 30 min | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GDSL-like Lipase/Acylhydrolase family protein | | Authors: | Williams, A.H, Gomperts Boneca, I. | | Deposit date: | 2013-04-11 | | Release date: | 2014-09-03 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Visualization of a substrate-induced productive conformation of the catalytic triad of the Neisseria meningitidis peptidoglycan O-acetylesterase reveals mechanistic conservation in SGNH esterase family members.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2Y4U
 
 | |
3ULA
 
 | | Crystal structure of the TV3 mutant F63W-MD-2-Eritoran complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-O-DECYL-2-DEOXY-6-O-{2-DEOXY-3-O-[(3R)-3-METHOXYDECYL]-6-O-METHYL-2-[(11Z)-OCTADEC-11-ENOYLAMINO]-4-O-PHOSPHONO-BETA-D-GLUCOPYRANOSYL}-2-[(3-OXOTETRADECANOYL)AMINO]-1-O-PHOSPHONO-ALPHA-D-GLUCOPYRANOSE, Lymphocyte antigen 96, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
5XTR
 
 | |
2XVC
 
 | | Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division | | Descriptor: | CADMIUM ION, CDVA, SSO0911, ... | | Authors: | Samson, R.Y, Obita, T, Hodgson, B, Shaw, M.K, Chong, P.L, Williams, R.L, Bell, S.D. | | Deposit date: | 2010-10-25 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular and Structural Basis of Escrt-III Recruitment to Membranes During Archaeal Cell Division.
Mol.Cell, 41, 2011
|
|
2XUB
 
 | | Human RPC62 subunit structure | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC3 | | Authors: | Lefevre, S, Legrand, P, Fribourg, S. | | Deposit date: | 2010-10-18 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Function Analysis of Hrpc62 Provides Insights Into RNA Polymerase III Transcription
Nat.Struct.Mol.Biol., 18, 2011
|
|
3V1A
 
 | | Crystal structure of de novo designed MID1-apo1 | | Descriptor: | Computational design, MID1-apo1 | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1E
 
 | | Crystal structure of de novo designed MID1-zinc H12E mutant | | Descriptor: | Computational design, MID1-zinc H12E mutant, ZINC ION | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.073 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
2YPG
 
 | | Haemagglutinin of 1968 Human H3N2 Virus in Complex with Human Receptor Analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, Xiong, X, Haire, L.F, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
