1QK5
 
 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE WITH XMP, PYROPHOSPHATE AND TWO MG2+ IONS | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Heroux, A, White, E.L, Ross, L.J, Davis, R.L, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase with Xmp, Pyrophosphate and Two Mg2+ Ions Bound: Insights Into the Catalytic Mechanism
Biochemistry, 38, 1999
|
|
1QK3
 
 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE GMP COMPLEX | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Heroux, A, White, E.L, Ross, L.J, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase Gmp and -Imp Complexes: Comparison of Purine Binding Interactions with the Xmp Complex
Biochemistry, 38, 1999
|
|
1QK4
 
 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE IMP COMPLEX | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, INOSINIC ACID | | Authors: | Heroux, A, White, E.L, Ross, L.J, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase Gmp and -Imp Complexes: Comparison of Purine Binding Interactions with the Xmp Complex
Biochemistry, 38, 1999
|
|
1BYR
 
 | |
1BYS
 
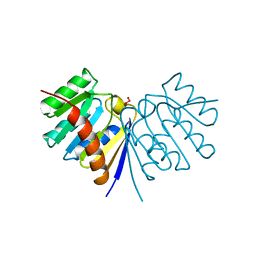 | |
1BYP
 
 | | E43K,D44K DOUBLE MUTANT PLASTOCYANIN FROM SILENE | | Descriptor: | COPPER (II) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Sugawara, H, Inoue, T, Li, C, Gotowda, M, Hibino, T, Takabe, T, Kai, Y. | | Deposit date: | 1998-10-19 | | Release date: | 1999-10-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene.
J.Biochem.(Tokyo), 125, 1999
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2BOS
 
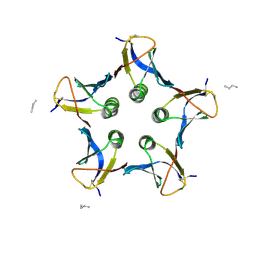 | | A MUTANT SHIGA-LIKE TOXIN IIE BOUND TO ITS RECEPTOR | | Descriptor: | N-BUTANE, PROTEIN (SHIGA-LIKE TOXIN IIE B SUBUNIT), alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Ling, H, Boodhoo, A, Armstrong, G.D, Clark, C.G, Brunton, J.L, Read, R.J. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A mutant Shiga-like toxin IIe bound to its receptor Gb(3): structure of a group II Shiga-like toxin with altered binding specificity.
Structure Fold.Des., 8, 2000
|
|
1CWQ
 
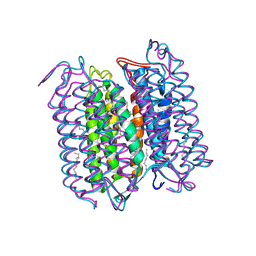 | | M INTERMEDIATE STRUCTURE OF THE WILD TYPE BACTERIORHODOPSIN IN COMBINATION WITH THE GROUND STATE STRUCTURE | | Descriptor: | BACTERIORHODOPSIN ("M" STATE INTERMEDIATE IN COMBINATION WITH GROUND STATE), HEXANE, N-OCTANE, ... | | Authors: | Sass, H.J, Berendzen, J, Neff, D, Gessenich, R, Ormos, P, Bueldt, G. | | Deposit date: | 1999-08-26 | | Release date: | 1999-10-20 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural alterations for proton translocation in the M state of wild-type bacteriorhodopsin.
Nature, 406, 2000
|
|
1C8S
 
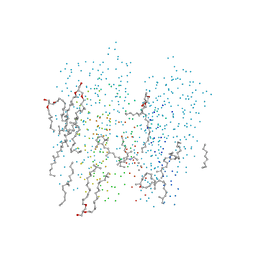 | | BACTERIORHODOPSIN D96N LATE M STATE INTERMEDIATE | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN ("M" STATE INTERMEDIATE), ... | | Authors: | Luecke, H. | | Deposit date: | 1999-07-29 | | Release date: | 1999-10-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution.
Science, 286, 1999
|
|
1C8R
 
 | | BACTERIORHODOPSIN D96N BR STATE AT 2.0 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, PROTEIN (BACTERIORHODOPSIN), ... | | Authors: | Luecke, H. | | Deposit date: | 1999-07-29 | | Release date: | 1999-10-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution.
Science, 286, 1999
|
|
1D5D
 
 | |
1D5H
 
 | | Rnase s(f8a). mutant ribonucleasE S. | | Descriptor: | RNASE S, S PEPTIDE, SULFATE ION | | Authors: | Ratnaparkhi, G.S, Varadarajan, R. | | Deposit date: | 1999-10-07 | | Release date: | 1999-10-20 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thermodynamic and structural studies of cavity formation in proteins suggest that loss of packing interactions rather than the hydrophobic effect dominates the observed energetics.
Biochemistry, 39, 2000
|
|
1D5E
 
 | |
1COU
 
 | |
1QP2
 
 | | SOLUTION STRUCTURE OF PHOTOSYSTEM I ACCESSORY PROTEIN E FROM THE CYANOBACTERIUM NOSTOC SP. STRAIN PCC 8009 | | Descriptor: | PROTEIN (PSAE PROTEIN) | | Authors: | Mayer, K.L, Shen, G, Bryant, D.A, Lecomte, J.T.J, Falzone, C.J. | | Deposit date: | 1999-05-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of photosystem I accessory protein E from the cyanobacterium Nostoc sp. strain PCC 8009.
Biochemistry, 38, 1999
|
|
1PVX
 
 | | DO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.5 | | Descriptor: | PROTEIN (ENDO-1,4-BETA-XYLANASE) | | Authors: | Rajeshkumar, P, Eswaramoorthy, S, Vithayathil, P.J, Viswamitra, M.A. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The tertiary structure at 1.59 A resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti bainier.
J.Mol.Biol., 295, 2000
|
|
1QB2
 
 | | CRYSTAL STRUCTURE OF THE CONSERVED SUBDOMAIN OF HUMAN PROTEIN SRP54M AT 2.1A RESOLUTION: EVIDENCE FOR THE MECHANISM OF SIGNAL PEPTIDE BINDING | | Descriptor: | HUMAN SIGNAL RECOGNITION PARTICLE 54 KD PROTEIN | | Authors: | Clemons Jr, W.M, Gowda, K, Black, S.D, Zwieb, C, Ramakrishnan, V. | | Deposit date: | 1999-04-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 A resolution: evidence for the mechanism of signal peptide binding.
J.Mol.Biol., 292, 1999
|
|
1QBH
 
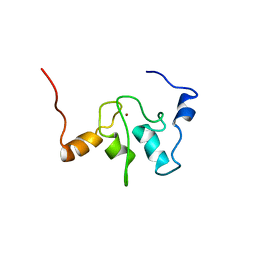 | | SOLUTION STRUCTURE OF A BACULOVIRAL INHIBITOR OF APOPTOSIS (IAP) REPEAT | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN (2MIHB/C-IAP-1), ZINC ION | | Authors: | Hinds, M.G, Norton, R.S, Vaux, D.L, Day, C.L. | | Deposit date: | 1999-04-20 | | Release date: | 1999-10-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat.
Nat.Struct.Biol., 6, 1999
|
|
1QP3
 
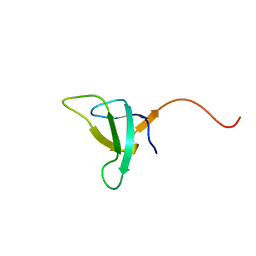 | | SOLUTION STRUCTURE OF PHOTOSYSTEM I ACCESSORY PROTEIN E FROM THE CYANOBACTERIUM NOSTOC SP. STRAIN PCC 8009 | | Descriptor: | PROTEIN (PSAE PROTEIN) | | Authors: | Mayer, K.L, Shen, G, Bryant, D.A, Lecomte, J.T.J, Falzone, C.J. | | Deposit date: | 1999-05-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of photosystem I accessory protein E from the cyanobacterium Nostoc sp. strain PCC 8009.
Biochemistry, 38, 1999
|
|
1BYL
 
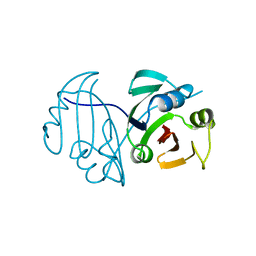 | |
1BX3
 
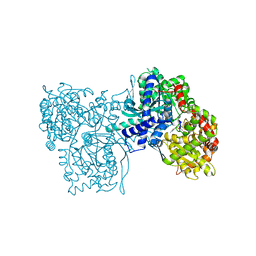 | | EFFECTS OF COMMONLY USED CRYOPROTECTANTS ON GLYCOGEN PHOSPHORYLASE ACTIVITY AND STRUCTURE | | Descriptor: | PROTEIN (GLYCOGEN PHOSPHORYLASE B), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tsitsanou, K.E, Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Gregoriou, M, Watson, K.A, Johnson, L.N, Fleet, G.W.J. | | Deposit date: | 1999-10-13 | | Release date: | 1999-10-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of commonly used cryoprotectants on glycogen phosphorylase activity and structure.
Protein Sci., 8, 1999
|
|
1D6O
 
 | | NATIVE FKBP | | Descriptor: | AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), SULFATE ION | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-15 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7H
 
 | | FKBP COMPLEXED WITH DMSO | | Descriptor: | AMMONIUM ION, DIMETHYL SULFOXIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7J
 
 | | FKBP COMPLEXED WITH 4-HYDROXY-2-BUTANONE | | Descriptor: | 4-HYDROXY-2-BUTANONE, AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
