7VIG
 
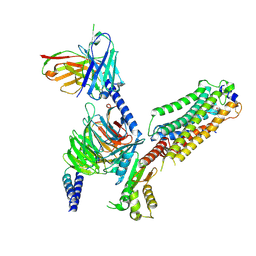 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2H68
 
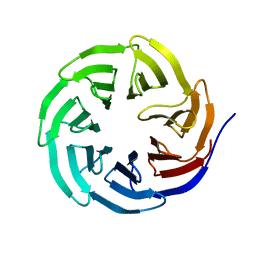 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-30 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H6K
 
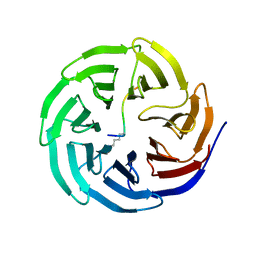 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H9L
 
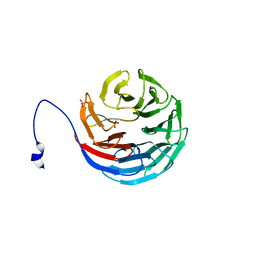 | | WDR5delta23 | | Descriptor: | SULFATE ION, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
5MES
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 29 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[[4-chloranyl-3-(2-phenylethyl)phenyl]methyl]-13-[(4-chlorophenyl)methyl]-8-methyl-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20(25),21,23-triene-3,7,15,26-tetrone, Heavy Chain, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
7UY1
 
 | | HUMAN PRMT5:MEP50 COMPLEX WITH MTA and Fragment 5 Bound | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-1,5-naphthyridin-2-amine, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Gunn, R.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7VIE
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VIF
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with (S)-FTY720-P | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5MFT
 
 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-1-bromo-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
7VIH
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4P8Q
 
 | | Crystal Structure of Human Insulin Regulated Aminopeptidase with Alanine in Active Site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucyl-cystinyl aminopeptidase, ... | | Authors: | Hermans, S.J, Ascher, D.B, Hancock, N.C, Holien, J.K, Michell, B, Morton, C.J, Parker, M.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides.
Protein Sci., 24, 2015
|
|
1MXE
 
 | | Structure of the Complex of Calmodulin with the Target Sequence of CaMKI | | Descriptor: | CALCIUM ION, Calmodulin, Target Sequence of rat Calmodulin-Dependent Protein Kinase I | | Authors: | Clapperton, J.A, Martin, S.R, Smerdon, S.J, Gamblin, S.J, Bayley, P.M. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Complex
of Calmodulin with the Target
Sequence of Calmodulin-Dependent
Protein Kinase I: Studies of the
Kinase Activation Mechanism
Biochemistry, 41, 2002
|
|
4TQO
 
 | |
7BCY
 
 | |
7BED
 
 | |
5MRB
 
 | | Crystal structure of human Mps1 (TTK) in complex with Cpd-5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dual specificity protein kinase TTK, ... | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-12-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Understanding inhibitor resistance in Mps1 kinase through novel biophysical assays and structures.
J. Biol. Chem., 292, 2017
|
|
5MFR
 
 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
4TQC
 
 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2S)-3-(4-amino-3-nitrophenyl)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinyl}propanoic acid, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2G99
 
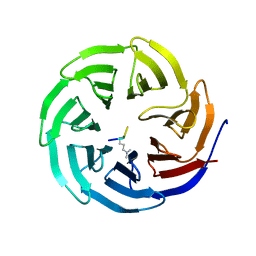 | |
5U4I
 
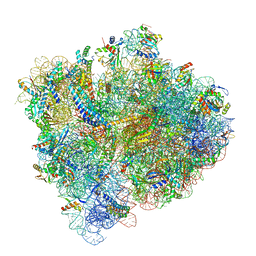 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
2H14
 
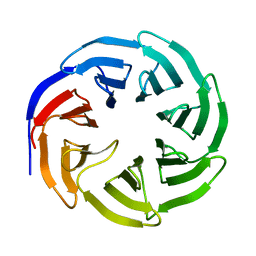 | |
7WUI
 
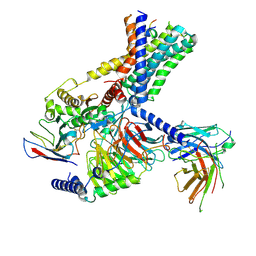 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
4Q4E
 
 | | Crystal structure of E.coli aminopeptidase N in complex with actinonin | | Descriptor: | ACTINONIN, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
7WU3
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7X9B
 
 | | Cryo-EM structure of neuropeptide Y Y2 receptor in complex with NPY and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Sci Adv, 8, 2022
|
|
