1CVJ
 
 | | X-RAY CRYSTAL STRUCTURE OF THE POLY(A)-BINDING PROTEIN IN COMPLEX WITH POLYADENYLATE RNA | | Descriptor: | 5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', ADENOSINE MONOPHOSPHATE, POLYADENYLATE BINDING PROTEIN 1 | | Authors: | Deo, R.C, Bonanno, J.B, Sonenberg, N, Burley, S.K. | | Deposit date: | 1999-08-23 | | Release date: | 1999-10-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of polyadenylate RNA by the poly(A)-binding protein.
Cell(Cambridge,Mass.), 98, 1999
|
|
1QLQ
 
 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
1B56
 
 | | HUMAN RECOMBINANT EPIDERMAL FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID BINDING PROTEIN, PALMITIC ACID | | Authors: | Van Tilbeurgh, H, Hohoff, C, Borchers, T, Spener, F. | | Deposit date: | 1999-01-12 | | Release date: | 1999-10-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein.
Biochemistry, 38, 1999
|
|
1B8L
 
 | | Calcium-bound D51A/E101D/F102W Triple Mutant of Beta Carp Parvalbumin | | Descriptor: | CALCIUM ION, CARBONATE ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-02-01 | | Release date: | 1999-10-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
1QEX
 
 | | BACTERIOPHAGE T4 GENE PRODUCT 9 (GP9), THE TRIGGER OF TAIL CONTRACTION AND THE LONG TAIL FIBERS CONNECTOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PROTEIN (BACTERIOPHAGE T4 GENE PRODUCT 9 (GP9)) | | Authors: | Kostyuchenko, V.A, Navruzbekov, G.A, Kurochkina, L.P, Strelkov, S.V, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 1999-03-30 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of bacteriophage T4 gene product 9: the trigger for tail contraction.
Structure Fold.Des., 7, 1999
|
|
1OTH
 
 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE TRANSCARBAMOYLASE COMPLEXED WITH N-PHOSPHONACETYL-L-ORNITHINE | | Descriptor: | N-(PHOSPHONOACETYL)-L-ORNITHINE, PROTEIN (ORNITHINE TRANSCARBAMOYLASE) | | Authors: | Shi, D, Morizono, H, Ha, Y, Aoyagi, M, Tuchman, N, Allewell, N.M. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85-A resolution crystal structure of human ornithine transcarbamoylase complexed with N-phosphonacetyl-L-ornithine. Catalytic mechanism and correlation with inherited deficiency.
J.Biol.Chem., 273, 1998
|
|
1C9N
 
 | |
1COZ
 
 | | CTP:GLYCEROL-3-PHOSPHATE CYTIDYLYLTRANSFERASE FROM BACILLUS SUBTILIS | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, PROTEIN (GLYCEROL-3-PHOSPHATE CYTIDYLYLTRANSFERASE) | | Authors: | Weber, C.H, Park, Y.S, Sanker, S, Kent, C, Ludwig, M.L. | | Deposit date: | 1999-05-29 | | Release date: | 1999-10-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A prototypical cytidylyltransferase: CTP:glycerol-3-phosphate cytidylyltransferase from bacillus subtilis.
Structure Fold.Des., 7, 1999
|
|
1BXN
 
 | | THE CRYSTAL STRUCTURE OF RUBISCO FROM ALCALIGENES EUTROPHUS TO 2.7 ANGSTROMS. | | Descriptor: | PHOSPHATE ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN), PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN) | | Authors: | Hansen, S, Vollan, V.B, Hough, E, Andersen, K. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of rubisco from Alcaligenes eutrophus reveals a novel central eight-stranded beta-barrel formed by beta-strands from four subunits.
J.Mol.Biol., 288, 1999
|
|
1C9J
 
 | |
1C9M
 
 | | BACILLUS LENTUS SUBTILSIN (SER 87) N76D/S103A/V104I | | Descriptor: | CALCIUM ION, SERINE PROTEASE, SULFATE ION | | Authors: | Bott, R. | | Deposit date: | 1999-08-02 | | Release date: | 1999-10-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Engineered Bacillus lentus subtilisins having altered flexibility.
J.Mol.Biol., 292, 1999
|
|
1CLI
 
 | | X-RAY CRYSTAL STRUCTURE OF AMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE (PURM), FROM THE E. COLI PURINE BIOSYNTHETIC PATHWAY, AT 2.5 A RESOLUTION | | Descriptor: | PROTEIN (PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE), SULFATE ION | | Authors: | Li, C, Kappock, T.J, Stubbe, J, Weaver, T.M, Ealick, S.E. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution.
Structure Fold.Des., 7, 1999
|
|
2PAH
 
 | |
1D1R
 
 | |
1D1L
 
 | | CRYSTAL STRUCTURE OF CRO-F58W MUTANT | | Descriptor: | LAMBDA CRO REPRESSOR, SULFATE ION | | Authors: | Rupert, P.B, Mollah, A.K, Mossing, M.C, Matthews, B.W. | | Deposit date: | 1999-09-17 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for enhanced stability and reduced DNA binding seen in engineered second-generation Cro monomers and dimers.
J.Mol.Biol., 296, 2000
|
|
1D2C
 
 | | METHYLTRANSFERASE | | Descriptor: | PROTEIN (GLYCINE N-METHYLTRANSFERASE) | | Authors: | Huang, Y, Takusagawa, F. | | Deposit date: | 1999-09-23 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1QHU
 
 | | MAMMALIAN BLOOD SERUM HAEMOPEXIN DEGLYCOSYLATED AND IN COMPLEX WITH ITS LIGAND HAEM | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PROTEIN (HEMOPEXIN), ... | | Authors: | Paoli, M, Baker, H.M, Morgan, W.T, Smith, A, Baker, E.N. | | Deposit date: | 1999-05-27 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two beta-propeller domains.
Nat.Struct.Biol., 6, 1999
|
|
1CUN
 
 | | CRYSTAL STRUCTURE OF REPEATS 16 AND 17 OF CHICKEN BRAIN ALPHA SPECTRIN | | Descriptor: | PROTEIN (ALPHA SPECTRIN) | | Authors: | Grum, V.L, Li, D, MacDonald, R.I, Mondragon, A. | | Deposit date: | 1999-08-20 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two repeats of spectrin suggest models of flexibility.
Cell(Cambridge,Mass.), 98, 1999
|
|
1QJ9
 
 | |
1TC1
 
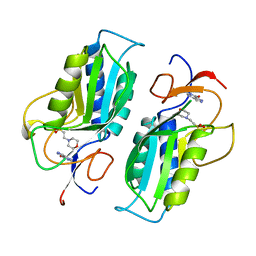 | | A 1.4 ANGSTROM CRYSTAL STRUCTURE FOR THE HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE OF TRYPANOSOMA CRUZI | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMYCIN B, PROTEIN (HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE) | | Authors: | Focia, P.J, Craig III, S.P, Nieves-Alicea, R, Fletterick, R.J, Eakin, A.E. | | Deposit date: | 1998-09-30 | | Release date: | 1999-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A 1.4 A crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi.
Biochemistry, 37, 1998
|
|
2THI
 
 | |
3HDH
 
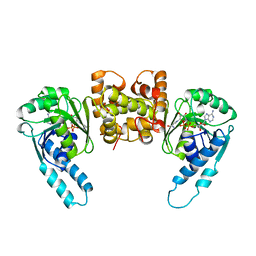 | | PIG HEART SHORT CHAIN L-3-HYDROXYACYL COA DEHYDROGENASE REVISITED: SEQUENCE ANALYSIS AND CRYSTAL STRUCTURE DETERMINATION | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (L-3-HYDROXYACYL COA DEHYDROGENASE) | | Authors: | Barycki, J.J, O'Brien, L.K, Birktoft, J.J, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 1999-04-13 | | Release date: | 1999-10-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pig heart short chain L-3-hydroxyacyl-CoA dehydrogenase revisited: sequence analysis and crystal structure determination.
Protein Sci., 8, 1999
|
|
1B7G
 
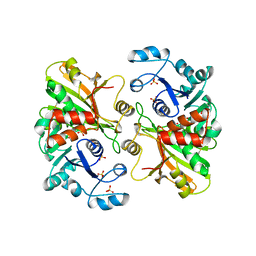 | | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE | | Descriptor: | PROTEIN (GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Isupov, M.N, Littlechild, J.A. | | Deposit date: | 1999-01-22 | | Release date: | 1999-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
J.Mol.Biol., 291, 1999
|
|
1C1F
 
 | | LIGAND-FREE CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I) | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1CXZ
 
 | | CRYSTAL STRUCTURE OF HUMAN RHOA COMPLEXED WITH THE EFFECTOR DOMAIN OF THE PROTEIN KINASE PKN/PRK1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PROTEIN (HIS-TAGGED TRANSFORMING PROTEIN RHOA(0-181)), ... | | Authors: | Maesaki, R, Ihara, K, Shimizu, T, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-08-31 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1.
Mol.Cell, 4, 1999
|
|
