6EGF
 
 | |
4ZFB
 
 | | Cytochrome P450 pentamutant from BM3 bound to Palmitic Acid | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
4ZFA
 
 | | Cytochrome P450 wild type from BM3 with bound PEG | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.765 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
5XB0
 
 | |
4ZF6
 
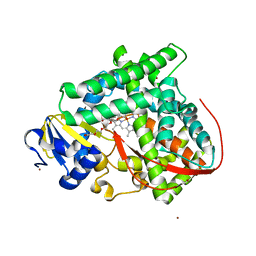 | | Cytochrome P450 pentamutant from BM3 with bound PEG | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
6PXN
 
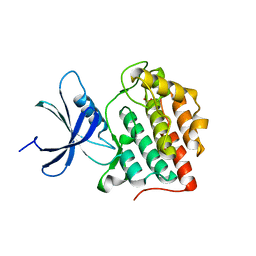 | |
6TT0
 
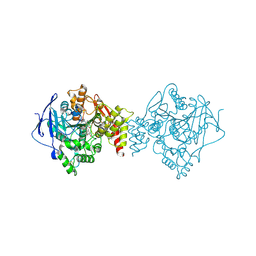 | | Crystal structure of a potent and reversible dual binding site Acetylcholinesterase chiral inhibitor | | Descriptor: | (1~{R},3~{S})-~{N}-(6,7-dimethoxy-2-oxidanylidene-chromen-3-yl)-3-[(phenylmethyl)amino]cyclohexane-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | de la Mora, E, Mangiatordi, G.F, Belviso, B.D, Caliandro, R, Colletier, J.P, Catto, M. | | Deposit date: | 2019-12-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.80003023 Å) | | Cite: | Chiral Separation, X-ray Structure, and Biological Evaluation of a Potent and Reversible Dual Binding Site AChE Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
5N4O
 
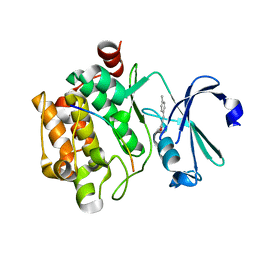 | |
5N52
 
 | |
6U41
 
 | | 1.7 angstrom structure of a pathogenic human Syt 1 C2B (D304G) | | Descriptor: | SULFATE ION, Synaptotagmin-1 | | Authors: | Dominguez, M.J, Bradberry, M.M, Chapman, E.R, Sutton, R.B. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for Synaptotagmin-1-Associated Neurodevelopmental Disorder.
Neuron, 107, 2020
|
|
5K8K
 
 | | Structure of the Haemophilus influenzae LpxH-lipid X complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, ACETATE ION, GLYCEROL, ... | | Authors: | Cho, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the essential Haemophilus influenzae UDP-diacylglucosamine pyrophosphohydrolase LpxH in lipid A biosynthesis.
Nat Microbiol, 1, 2016
|
|
7EO3
 
 | | X-ray structure analysis of beita-1,3-glucanase | | Descriptor: | 1,3-beta-glucanase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION | | Authors: | Wan, Q, Feng, J, Xu, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.141 Å) | | Cite: | Identification and structural analysis of a thermophilic beta-1,3-glucanase from compost
Bioresour Bioprocess, 8, 2021
|
|
6B5H
 
 | |
5GUV
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - R1279D | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Ye, F, Chen, S.J. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5N5M
 
 | |
5N9Z
 
 | | Rubisco from Thalassiosira hyalina | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Andersson, I, Valegard, K. | | Deposit date: | 2017-02-27 | | Release date: | 2018-03-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural and functional analyses of Rubisco from arctic diatom species reveal unusual posttranslational modifications.
J. Biol. Chem., 293, 2018
|
|
5KA6
 
 | |
7VC7
 
 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kojima, K, Sunagawa, N, Igarashi, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
5N50
 
 | |
5KQL
 
 | |
9B3S
 
 | |
6KVI
 
 | | Crystal structure of UDP-SrUGT76G1 | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
5YEQ
 
 | | The structure of Sac-KARI protein | | Descriptor: | 1,2-ETHANEDIOL, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Ko, T.P, Chen, C.Y, Lin, K.F, Lin, B.L, Huang, C.H, Chiang, C.H, Horng, J.C, Tsai, M.D. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NADH/NADPH bi-cofactor-utilizing and thermoactive ketol-acid reductoisomerase from Sulfolobus acidocaldarius
Sci Rep, 8, 2018
|
|
6UAQ
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) | | Descriptor: | GLYCEROL, Glyco_hydro_cc domain-containing protein, SODIUM ION | | Authors: | Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6B5I
 
 | |
