3Q6M
 
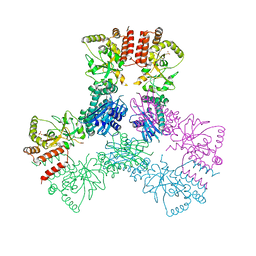 | | Crystal Structure of Human MC-HSP90 in C2221 Space Group | | Descriptor: | Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Lee, C.C, Lin, T.W, Ko, T.P, Wang, A.H.-J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The hexameric structures of human heat shock protein 90
Plos One, 6, 2011
|
|
9EPL
 
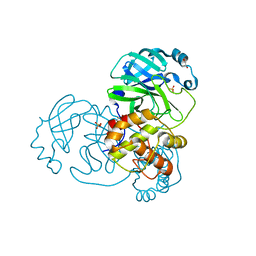 | | Mpro from SARS-CoV-2 with 298Q mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Non-structural protein 11, ... | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-18 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
8VE4
 
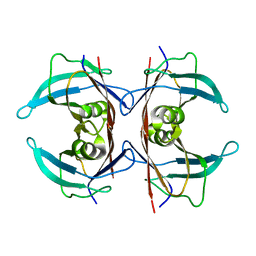 | | Unliganded human transthyretin in the frayed conformation | | Descriptor: | Transthyretin | | Authors: | Basanta, B, Nugroho, K, Yan, N, Kline, G.M, Tsai, F.J, Wu, M, Kelly, J.W, Lander, G.C. | | Deposit date: | 2023-12-18 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The conformational landscape of human transthyretin revealed by cryo-EM
To Be Published
|
|
8TZ3
 
 | |
8VEO
 
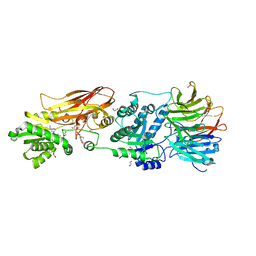 | | Crystal structure of PRMT5:MEP50 in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VXY
 
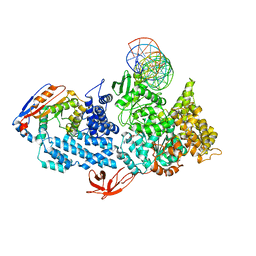 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | Authors: | Tuck, O.T, Hu, J.J, Doudna, J.A. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8TOR
 
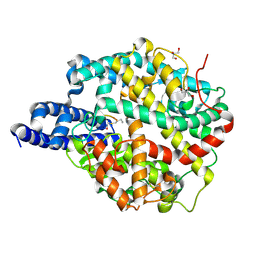 | | ACE2-peptide 2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8UP3
 
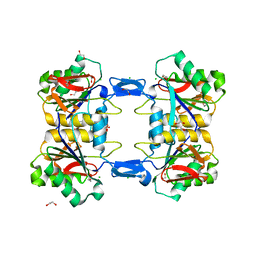 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F) | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, Asparaginase, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8VX9
 
 | |
8TU9
 
 | | Cryo-EM structure of HGSNAT-acetyl-CoA complex at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-acetyltransferase,Isoform 2 of Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Navratna, V, Kumar, A, Mosalaganti, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human heparan-alpha-glucosaminide N-acetyltransferase (HGSNAT)
eLife, 13, 2024
|
|
6S7C
 
 | | Crystal structure of CARM1 in complex with inhibitor UM079 | | Descriptor: | 1-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]propyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
8UPC
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K158M) | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
6RG6
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[3-(6-azanyl-9~{H}-purin-8-yl)prop-2-ynoxymethyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
8VGZ
 
 | |
6RG7
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-(6-azanyl-9~{H}-purin-8-yl)prop-2-ynyl-methyl-amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
8W43
 
 | | X-ray crystal structure of V30M-TTR in complex with piceatannol | | Descriptor: | PICEATANNOL, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W45
 
 | | X-ray crystal structure of V30M-TTR in complex with pinostilbene | | Descriptor: | 3-[(E)-2-(4-hydroxyphenyl)ethenyl]-5-methoxy-phenol, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8ZM2
 
 | | Structure of human pyruvate dehydrogenase kinase 2 complexed with compound 16 | | Descriptor: | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial, methyl (9~{R})-9-oxidanyl-9-(trifluoromethyl)fluorene-4-carboxylate | | Authors: | Akai, S, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-22 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design and synthesis of novel fluorene derivatives as inhibitors of pyruvate dehydrogenase kinase.
Bioorg.Med.Chem.Lett., 109, 2024
|
|
8XS3
 
 | | Structure of MPXV B6 and D68 fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D68_heavy chain, ... | | Authors: | wu, L.L, Sun, J.Q. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two noncompeting human neutralizing antibodies targeting MPXV B6 show protective effects against orthopoxvirus infections.
Nat Commun, 15, 2024
|
|
8VXA
 
 | |
6RGA
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2-(aminomethyl)-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
8W47
 
 | | X-ray crystal structure of V30M-TTR in complex with isorhapontigenin | | Descriptor: | 5-[(~{E})-2-(3-methoxy-4-oxidanyl-phenyl)ethenyl]benzene-1,3-diol, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
6RGB
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynylamino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
8UYF
 
 | | Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | Serine/threonine-protein kinase Pink1, mitochondrial | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
8V9A
 
 | | GII.NA1 Loreto 1257 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1 | | Authors: | Kher, G, Kim, I, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 2024
|
|
