3CJP
 
 | | Crystal structure of an uncharacterized amidohydrolase CAC3332 from Clostridium acetobutylicum | | Descriptor: | Predicted amidohydrolase, dihydroorotase family, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U.A, Bonanno, J.B, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase CAC3332 from Clostridium acetobutylicum.
To be Published
|
|
4Q2P
 
 | | NHERF3 PDZ2 in Complex with a Phage-Derived Peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | | Authors: | Appleton, B.A, Wiesmann, C. | | Deposit date: | 2014-04-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural portrait of the PDZ domain family.
J.Mol.Biol., 426, 2014
|
|
3CK9
 
 | | B. thetaiotaomicron SusD with maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SusD, ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
3CKY
 
 | |
3V9J
 
 | |
3VWQ
 
 | | 6-aminohexanoate-dimer hydrolase S112A/G181D/R187A/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VAI
 
 | | Structure of U2AF65 variant with BrU3C5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*UP*CP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Frato, K.H, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
4Q4L
 
 | |
3VAS
 
 | | Adenosine kinase from Schistosoma mansoni in complex with adenosine in occluded loop conformation | | Descriptor: | ADENOSINE, CHLORIDE ION, Putative adenosine kinase | | Authors: | Romanello, L, Bachega, F.R, Garatt, R.C, DeMarco, R, Brandao-neto, J, Pereira, H.M. | | Deposit date: | 2011-12-29 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Adenosine kinase from Schistosoma mansoni: structural basis for the differential incorporation of nucleoside analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3COU
 
 | | Crystal structure of human Nudix motif 16 (NUDT16) | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 16 | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Van Den Berg, S, Welin, M, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Nudix motif 16 (NUDT16).
To be Published
|
|
3CQH
 
 | |
4Q5G
 
 | | Crystal Structure of mouse Serum Amyloid A3 | | Descriptor: | Serum amyloid A-3 protein | | Authors: | Derebe, M.G, Hooper, L.V. | | Deposit date: | 2014-04-16 | | Release date: | 2014-09-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Biochemical and structural analysis reveals a retinol binding function for serum amyloid A proteins
elife, 2014
|
|
3VDD
 
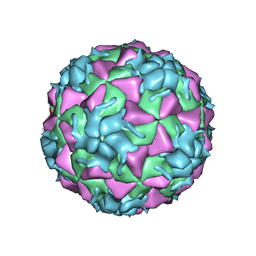 | | Structure of HRV2 capsid complexed with antiviral compound BTA798 | | Descriptor: | 3-ethoxy-6-{2-[1-(6-methylpyridazin-3-yl)piperidin-4-yl]ethoxy}-1,2-benzoxazole, Protein VP1, Protein VP2, ... | | Authors: | Morton, C.J, Feil, S.C, Parker, M.W. | | Deposit date: | 2012-01-05 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An Orally Available 3-Ethoxybenzisoxazole Capsid Binder with Clinical Activity against Human Rhinovirus.
ACS Med Chem Lett, 3, 2012
|
|
3CR4
 
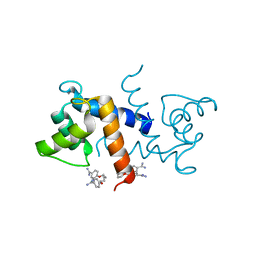 | | X-ray structure of bovine Pnt,Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3W11
 
 | |
3VE2
 
 | |
3VES
 
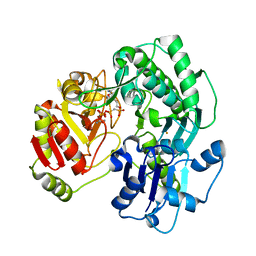 | | Crystal structure of the O-carbamoyltransferase TobZ in complex with AMPCPP and carbamoyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FE (II) ION, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4Q6E
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[3-(3,5-Dimethyl-1H-pyrazol-1-yl)-3-oxopropyl]amino}benzene-1-sulfonamide | | Descriptor: | 4-{[3-(3,5-dimethyl-1H-pyrazol-1-yl)-3-oxopropyl]amino}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 4-Amino-substituted Benzenesulfonamides as Inhibitors of Human Carbonic Anhydrases.
Molecules, 19, 2014
|
|
4Q7T
 
 | |
3CY3
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V | | Descriptor: | (2S)-1,3-benzothiazol-2-yl{2-[(2-pyridin-3-ylethyl)amino]pyrimidin-4-yl}ethanenitrile, 1,2-ETHANEDIOL, Pimtide peptide, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V.
To be Published
|
|
3CU5
 
 | | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg | | Descriptor: | Two component transcriptional regulator, AraC family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg.
To be Published
|
|
3CUJ
 
 | |
4Q7R
 
 | |
3VGM
 
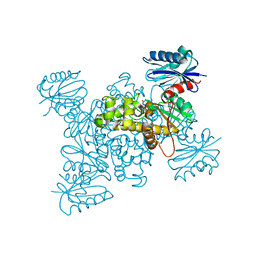 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose | | Descriptor: | Glucokinase, POTASSIUM ION, ZINC ION, ... | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
3V9A
 
 | |
