6U03
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6U00
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6TZO
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6TZM
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
3WOF
 
 | | Crystal structure of P23-45 gp39 (6-132) bound to Thermus thermophilus RNA polymerase beta-flap domain | | Descriptor: | DNA-directed RNA polymerase subunit beta, Putative uncharacterized protein | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.298 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
2VOD
 
 | | Crystal structure of N-terminal domains of Human La protein complexed with RNA oligomer AUAUUUU | | Descriptor: | 5'-R(*AP*UP*AP*UP*UP*UP*UP)-3', LUPUS LA PROTEIN | | Authors: | Kotik-Kogan, O, Valentine, E.R, Sanfelice, D, Conte, M.R, Curry, S. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3' Ends by the Human La Protein.
Structure, 16, 2008
|
|
2JC0
 
 | | CRYSTAL STRUCTURE OF HEPATITIS C VIRUS POLYMERASE IN COMPLEX WITH INHIBITOR SB655264 | | Descriptor: | (2S,4S,5R)-2-ISOBUTYL-5-(2-THIENYL)-1-[4-(TRIFLUOROMETHYL)BENZOYL]PYRROLIDINE-2,4-DICARBOXYLIC ACID, RNA-DEPENDENT RNA-POLYMERASE | | Authors: | Wonacott, A, Skarzynski, T, Singh, O.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of Novel Acyl Pyrrolidine Inhibitors of Hepatitis C Virus RNA-Dependent RNA Polymerase Leading to a Development Candidate.
J.Med.Chem., 50, 2007
|
|
2JC1
 
 | | CRYSTAL STRUCTURE OF HEPATITIS C VIRUS POLYMERASE IN COMPLEX WITH INHIBITOR SB698223 | | Descriptor: | (2S,4S,5R)-1-(4-TERT-BUTYLBENZOYL)-2-ISOBUTYL-5-(1,3-THIAZOL-2-YL)PYRROLIDINE-2,4-DICARBOXYLIC ACID, RNA-DEPENDENT RNA-POLYMERASE | | Authors: | Wonacott, A, Skarzynski, T, Singh, O.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of Novel Acyl Pyrrolidine Inhibitors of Hepatitis C Virus RNA-Dependent RNA Polymerase Leading to a Development Candidate.
J.Med.Chem., 50, 2007
|
|
8D35
 
 | |
2HFZ
 
 | |
6U05
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
2H1M
 
 | |
6CB3
 
 | |
6CC3
 
 | |
2I91
 
 | | 60kDa Ro autoantigen in complex with a fragment of misfolded RNA | | Descriptor: | 5'-R(*C*GP*GP*UP*AP*GP*GP*CP*UP*UP*UP*UP*CP*AP*A)-3', 5'-R(*GP*CP*CP*UP*AP*CP*CP*C)-3', 60 kDa SS-A/Ro ribonucleoprotein, ... | | Authors: | Reinisch, K.M, Stein, A.J. | | Deposit date: | 2006-09-04 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4KQ0
 
 | | Crystal structure of double-helical CGG-repetitive RNA 19mer complexed with RSS p19 | | Descriptor: | 5'-R(P*GP*GP*CP*GP*GP*CP*GP*GP*CP*GP*GP*CP*GP*GP*CP*GP*GP*CP*C)-3', RNA silencing suppressor p19, SULFATE ION | | Authors: | Cabo, A, Katorcha, E, Tamjar, J, Popov, A.N, Malinina, L. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into CNG-repetitive RNAs associated with human Trinucleotide Repeat Expansion Diseases (TREDs)
To be Published
|
|
3BX2
 
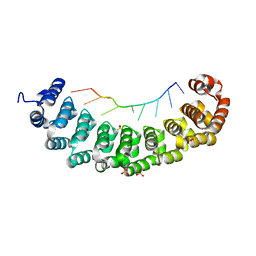 | | Puf4 RNA binding domain bound to HO endonuclease RNA 3' UTR recognition sequence | | Descriptor: | HO endonuclease 3' UTR binding sequence, Protein PUF4, SODIUM ION, ... | | Authors: | Miller, M.T, Higgin, J.J, Hall, T.M.T. | | Deposit date: | 2008-01-11 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p
Nat.Struct.Mol.Biol., 15, 2008
|
|
6CC1
 
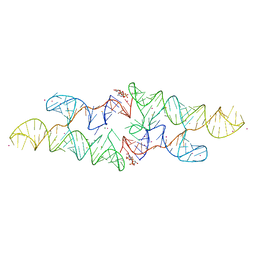 | |
2VOO
 
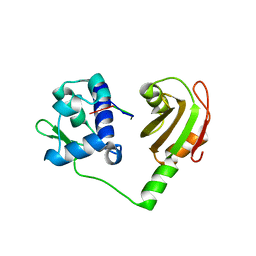 | | Crystal structure of N-terminal domains of Human La protein complexed with RNA oligomer UUUUUUUU | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP)-3', LUPUS LA PROTEIN | | Authors: | Kotik-Kogan, O, Valentine, E.R, Sanfelice, D, Conte, M.R, Curry, S. | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3' Ends by the Human La Protein.
Structure, 16, 2008
|
|
4KNQ
 
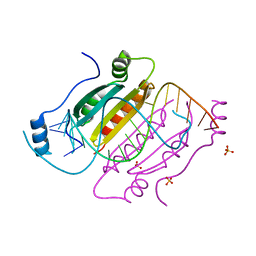 | |
2HCN
 
 | |
3C66
 
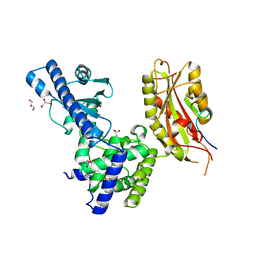 | | Yeast poly(A) polymerase in complex with Fip1 residues 80-105 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Poly(A) polymerase, ... | | Authors: | Bohm, A, Meinke, G. | | Deposit date: | 2008-02-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of yeast poly(A) polymerase in complex with a peptide from Fip1, an intrinsically disordered protein.
Biochemistry, 47, 2008
|
|
3CZ3
 
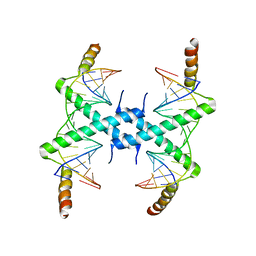 | | Crystal structure of Tomato Aspermy Virus 2b in complex with siRNA | | Descriptor: | Protein 2b, RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*A)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*G)-3') | | Authors: | Ma, J.B, Li, F, Ding, S.W, Patel, D.J. | | Deposit date: | 2008-04-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural Basis for siRNA Recognition by 2b, a Viral Suppressor of Non-Cell Autonomous RNA Silencing
To be Published
|
|
6IVU
 
 | |
8PM4
 
 | | Cryo-EM structure of the Cas12m-crRNA-target DNA complex | | Descriptor: | DNA oligoduplex, non-target strand, chain D, ... | | Authors: | Sasnauskas, G, Tamulaitiene, G, Karvelis, T, Bigelyte, G, Siksnys, V. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Innate programmable DNA binding by CRISPR-Cas12m effectors enable efficient base editing.
Nucleic Acids Res., 52, 2024
|
|
