8SNG
 
 | |
2WQR
 
 | | The high resolution crystal structure of IgE Fc | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, IG EPSILON CHAIN C REGION, ... | | Authors: | Dhaliwal, B, Sutton, B.J, Beavil, A.J. | | Deposit date: | 2009-08-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Changes in Ige Contribute to its Uniquely Slow Dissociation Rate from Receptor Fceri
Nat.Struct.Mol.Biol., 18, 2011
|
|
7QTB
 
 | | S1 nuclease from Aspergillus oryzae in complex with cytidine-5'-monophosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QTA
 
 | | S1 nuclease from Aspergillus oryzae in complex with uridine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QQJ
 
 | | Sucrose phosphorylase from Jeotgalibaca ciconiae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Sucrose phosphorylase | | Authors: | Ubiparip, Z, Capra, N, Rozeboom, H.J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2022-01-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sucrose phosphorylase from Jeotgalibaca ciconiae
To Be Published
|
|
8EWQ
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {N-[1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]benzamide}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWE
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-22 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWR
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWD
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-22 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWM
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methylidene]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[3,5-difluoro-2-(pyridin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
7R2V
 
 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
6FV2
 
 | |
6WTQ
 
 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-4-{[4-(1-propyl-1H-pyrazol-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}benzamide, ... | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79968476 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6KEO
 
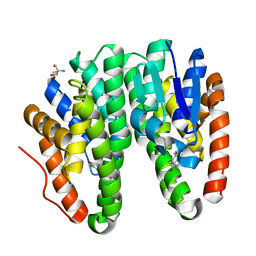 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in 17beta-estradiol-bound form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ESTRADIOL, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KEL
 
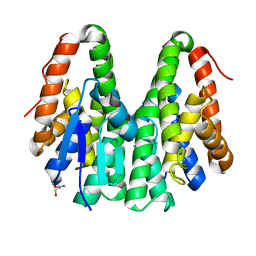 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in apo-form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6H40
 
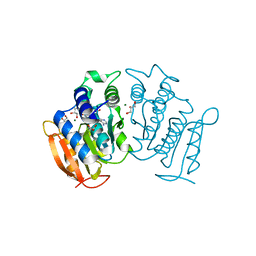 | | High resolution structure of MeT1 from Mycobacterium hassiacum in complex with 3-methoxy-1,2-propanediol. | | Descriptor: | (2~{S})-3-methoxypropane-1,2-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pereira, P.J.B, Ripoll-Rozada, J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.053 Å) | | Cite: | Biosynthesis of mycobacterial methylmannose polysaccharides requires a unique 1-O-methyltransferase specific for 3-O-methylated mannosides.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5TE6
 
 | |
3CJF
 
 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
3CJG
 
 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-methyl-N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
3HG0
 
 | | Crystal structure of a DARPin in complex with ORF49 from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein, Designed Ankyrin Repeat Protein (DARPin) 20 | | Authors: | Veesler, D, Dreier, B, Blangy, S, Lichiere, J, Tremblay, D, Moineau, S, Spinelli, S, Tegoni, M, Pluckthun, A, Campanacci, V, Cambillau, C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode.
J.Biol.Chem., 284, 2009
|
|
3LQR
 
 | | Structure of CED-4:CED-3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.896 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7EHF
 
 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmB1 | | Descriptor: | 1,2-ETHANEDIOL, 16S rRNA methyltransferase, CHLORIDE ION, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and Structural Characterization of Acquired 16S rRNA Methyltransferase NpmB1 Conferring Pan-Aminoglycoside Resistance.
Antimicrob.Agents Chemother., 65, 2021
|
|
5ENZ
 
 | | S. aureus MnaA-UDP co-structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chemical Genetic Analysis and Functional Characterization of Staphylococcal Wall Teichoic Acid 2-Epimerases Reveals Unconventional Antibiotic Drug Targets.
Plos Pathog., 12, 2016
|
|
4QGW
 
 | | Crystal sturcture of the R132K:R111L:L121D mutant of Cellular Retinoic Acid Binding ProteinII complexed with a synthetic ligand (Merocyanine) at 1.77 angstrom resolution | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | Authors: | Nosrati, M, Yapici, I, Geiger, J.H. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
