7V08
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a Spb1 D52A suppressor 3 strain | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
6FC3
 
 | |
4YKE
 
 | |
5DGV
 
 | | Complex of yeast 80S ribosome with hypusine-containing/non-modified eIF5A and/or a peptidyl-tRNA analog | | Descriptor: | 25S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-28 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
8OHD
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ8
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ0
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
5J8K
 
 | | Architecture of supercomplex I-III2 | | Descriptor: | COMPLEX I 13KDA/NDUFS6, COMPLEX I 15KDA/NDUFS5, COMPLEX I 18KDA/NDUFS6, ... | | Authors: | Letts, J.A, Fiedorczuk, K, Sazanov, L.A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-09-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The architecture of respiratory supercomplexes.
Nature, 537, 2016
|
|
4U1R
 
 | |
8D7R
 
 | |
7Z34
 
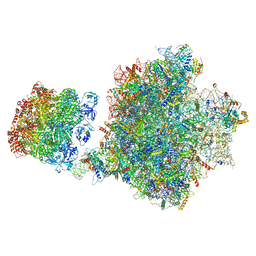 | | Structure of pre-60S particle bound to DRG1(AFG2). | | Descriptor: | 35S pre-ribosomal RNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Prattes, M, Grishkovskaya, I, Bergler, H, Haselbach, D. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-21 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4W8A
 
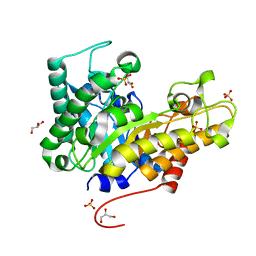 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | Exo-xyloglucanase, GLYCEROL, SULFATE ION | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
8BXY
 
 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2022-12-11 | | Release date: | 2023-04-12 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
8BY3
 
 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-12-11 | | Release date: | 2023-04-12 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
2W8S
 
 | | CRYSTAL STRUCTURE OF A catalytically promiscuous PHOSPHONATE MONOESTER HYDROLASE FROM Burkholderia caryophylli | | Descriptor: | FE (III) ION, GLYCEROL, PHOSPHONATE MONOESTER HYDROLASE, ... | | Authors: | Jonas, S, van Loo, B, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2009-01-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Efficient, Multiply Promiscuous Hydrolase in the Alkaline Phosphatase Superfamily.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4W68
 
 | |
2IPB
 
 | |
7T1W
 
 | |
7T1X
 
 | |
6CT3
 
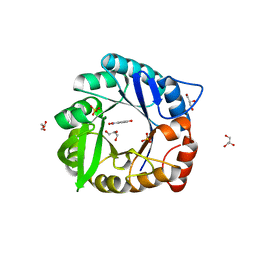 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 27 round 7-2 | | Descriptor: | 5-nitro-2-oxidanyl-benzenecarbonitrile, GLYCEROL, Kemp eliminase, ... | | Authors: | Jackson, C.J, Hong, N.-S, Carr, P.D. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
4W84
 
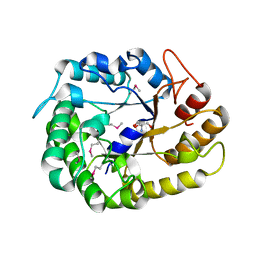 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
8CK3
 
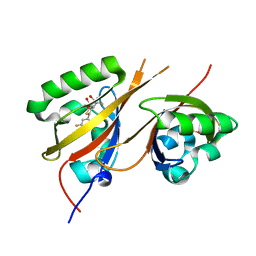 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (S)-1-(3,5-Difluoro-phenyl)-5,5-difluoro-3-methanesulfonyl-5,6-dihydro-4H-cyclopenta[c]thiophen-4-ol | | Descriptor: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-4,6-dihydrocyclopenta[c]thiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Lehmannn, M, Diehl, L. | | Deposit date: | 2023-02-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
4W6Z
 
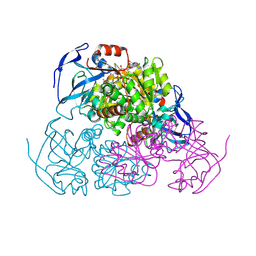 | | YEAST ALCOHOL DEHYDROGENASE I, SACCHAROMYCES CEREVISIAE FERMENTATIVE ENZYME | | Descriptor: | Alcohol dehydrogenase 1, NICOTINAMIDE-8-IODO-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | plapp, B.v, savarimuthu, b.r, ramaswamy, s. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Yeast alcohol dehydrogenase structure and catalysis.
Biochemistry, 53, 2014
|
|
8CK8
 
 | |
8CK4
 
 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (4S)-1-(3,5-difluorophenyl)-5,5-difluoro-3-methanesulfonyl-4,5,6,7-tetrahydro-2-benzothiophen-4-ol | | Descriptor: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-6,7-dihydro-4~{H}-2-benzothiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Musil, D. | | Deposit date: | 2023-02-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
