9GES
 
 | |
1MOT
 
 | | NMR Structure Of Extended Second Transmembrane Domain Of Glycine Receptor alpha1 Subunit in SDS Micelles | | Descriptor: | Glycine Receptor alpha-1 CHAIN | | Authors: | Yushmanov, V.E, Mandal, P.K, Liu, Z, Tang, P, Xu, Y. | | Deposit date: | 2002-09-09 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Backbone Dynamics of the Extended Second Transmembrane Domain of the Human Neuronal Glycine Receptor Alpha1 Subunit
Biochemistry, 42, 2003
|
|
1M1L
 
 | | Human Suppressor of Fused (N-terminal domain) | | Descriptor: | Suppressor of Fused | | Authors: | Merchant, M, Vajdos, F.F, Ultsch, M, Maun, H.R, Wendt, U, Cannon, J, Lazarus, R.A, de Vos, A.M, de Sauvage, F.J. | | Deposit date: | 2002-06-19 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Suppressor of fused regulates Gli activity through a dual binding mechanism
Mol.Cell.Biol., 24, 2004
|
|
9GG8
 
 | | Crystal structure of 14-3-3 sigma dC - C38N in complex with Tau pS214 peptide and covalent stabilizer 187 | | Descriptor: | 14-3-3 protein sigma, 4-(3,4-dihydro-2~{H}-quinoxalin-1-ylsulfonyl)benzaldehyde, Microtubule-associated protein tau | | Authors: | van den Oetelaar, M.C.M, van de Wijngaart, T.R, Brunsveld, L, Ottmann, C. | | Deposit date: | 2024-08-13 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Modulation of Tau Condensation with 14-3-3/Tau Molecular Glues
To Be Published
|
|
1M20
 
 | | Crystal Structure of F35Y Mutant of Trypsin-solubilized Fragment of Cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yao, P, Wu, J, Wang, Y.-H, Sun, B.-Y, Xia, Z.-X, Huang, Z.-X. | | Deposit date: | 2002-06-20 | | Release date: | 2002-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallography, CD and kinetic studies revealed the essence of the abnormal behaviors of the cytochrome b5 Phe35-->Tyr mutant.
Eur.J.Biochem., 269, 2002
|
|
1M25
 
 | | STRUCTURE OF SYNTHETIC 26-MER PEPTIDE CONTAINING 145-169 SHEEP PRION PROTEIN SEGMENT AND C-TERMINAL CYSTEINE IN TFE SOLUTION | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Megy, S, Bertho, G, Kozin, S.A, Coadou, G, Debey, P, Hoa, G.H, Girault, J.-P. | | Deposit date: | 2002-06-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Possible role of region 152-156 in the structural duality of a peptide fragment from sheep prion protein
Protein Sci., 13, 2004
|
|
9GE5
 
 | |
1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
9GJZ
 
 | | Ntaya methyltransferase (MTase) bound to AT-9010 (2'-methyl-2'-fluoro GTP) | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Boura, E, Krejcova, K. | | Deposit date: | 2024-08-23 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Ntaya methyltransferase (MTase) bound to AT-9010 (2'-methyl-2'-fluoro GTP)
To Be Published
|
|
7SCF
 
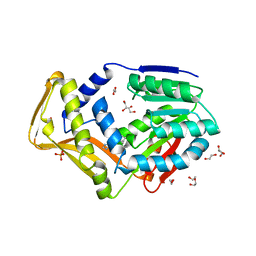 | | M. tb EgtD in complex with HD2 | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-(1H-pyrrol-1-yl)propanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sudasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-09-28 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Inhibitors of Mycobacterium tuberculosis EgtD target both substrate binding sites to limit hercynine production.
Sci Rep, 11, 2021
|
|
1MP7
 
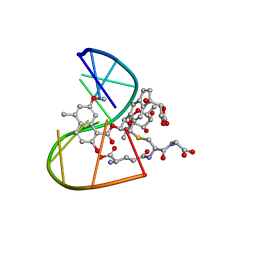 | | A Third Complex of Post-Activated Neocarzinostatin Chromophore with DNA. Bulge DNA Binding from the Minor Groove | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*AP*GP*AP*GP*C)-3', [(R)-4-((1,3-DIOXOLANE-2-OXY)-4-(S)-YL)-4-HYDROXY]-(R)-10-(2-METHYLAMINO-5-METHYL-2,6-DIDEOXYGALACTOPYRANOSYL-OXY)-(R)-11-(2-HYDROXY-5-METHYL-7-METHOXY-1-NAPHTHOYL-OXY)-(R)-12-S-GLUTATHIONYL-4,10,11,12-TETRAHYDROINDACENE | | Authors: | Kwon, Y, Xi, Z, Kappen, L.S, Goldberg, I.H, Gao, X. | | Deposit date: | 2002-09-11 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | New Complex of Post-Activated Neocarzinostatin Chromophore with DNA: Bulge DNA Binding from the Minor Groove
Biochemistry, 42, 2003
|
|
9GJY
 
 | |
1M2J
 
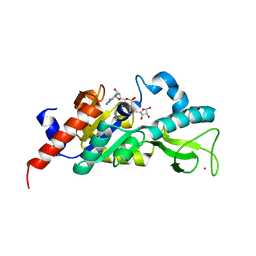 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
9GCE
 
 | | Ang-1 domain of the HupE/UreJ-2 protein from Rhodobacteraceae bacterium RbAng-1a with Cu bound | | Descriptor: | COPPER (II) ION, Membrane protein, ZINC ION | | Authors: | Franco Cairo, J.P.L, Correa, T.L.R, Offen, W.A, Nairn, A, Walton, J, Davies, G.J, Walton, P.H, Sweeney, S.T. | | Deposit date: | 2024-08-01 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Signal-strapping as a protein-sequence search method for the discovery of metalloproteins.
Nat Commun, 16, 2025
|
|
1M2X
 
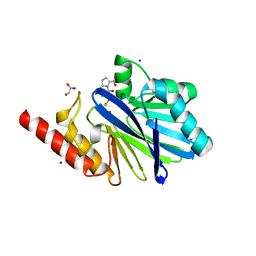 | |
1M3A
 
 | | Solution structure of a circular form of the truncated N-terminal SH3 domain from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
9GLD
 
 | |
9GJQ
 
 | |
1MQ5
 
 | |
1M3K
 
 | | biosynthetic thiolase, inactive C89A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-28 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M47
 
 | | Crystal Structure of Human Interleukin-2 | | Descriptor: | SULFATE ION, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, Wells, J.A, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Binding of small molecules to an adaptive protein-protein interface.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
9GNF
 
 | |
9GP3
 
 | | Crystal structure of CDK2-cyclin E1 bound by compound 11 (AZD8421) | | Descriptor: | (3~{S})-~{N}-ethyl-3-[[9-ethyl-2-[[(2~{R},3~{S})-2-oxidanylpentan-3-yl]amino]purin-6-yl]amino]pyrrolidine-1-sulfonamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Collie, G.W, Pflug, A. | | Deposit date: | 2024-09-06 | | Release date: | 2025-09-10 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of AZD8421: A Potent CDK2 Inhibitor with Selectivity Against Other CDK Family Members and the Human Kinome.
J.Med.Chem., 68, 2025
|
|
1M4C
 
 | | Crystal Structure of Human Interleukin-2 | | Descriptor: | interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MQJ
 
 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with willardiine at 1.65 angstroms resolution | | Descriptor: | 2-AMINO-3-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, ZINC ION, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
