6S0U
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
8EJN
 
 | | Structure of dehaloperoxidase A in complex with 2,4-dichlorophenol | | Descriptor: | 2,4-dichlorophenol, DIMETHYL SULFOXIDE, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Franzen, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Comparative study of the binding and activation of 2,4-dichlorophenol by dehaloperoxidase A and B.
J.Inorg.Biochem., 247, 2023
|
|
6RL5
 
 | |
8EHH
 
 | | Crystal structure of the class A extended-spectrum beta-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Ghiglione, B, Bonomo, R.A, Rodriguez, M.M, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Crystal structure of the class A extended-spectrum beta-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution.
Antimicrob.Agents Chemother., 68, 2024
|
|
6RWV
 
 | | Structure of apo-LmCpfC | | Descriptor: | Ferrochelatase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6386379 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
8ENJ
 
 | |
6RG2
 
 | | Photorhabdus laumondii lectin PLL2 in complex with 3-O-methyl-D-glucose | | Descriptor: | 3-O-methyl-alpha-D-glucopyranose, 3-O-methyl-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
6RGG
 
 | | Photorhabdus laumondii lectin PLL2 in complex with O-methylated PGL-1-derived disaccharide | | Descriptor: | 1,2-ETHANEDIOL, 3,6-O-dimethyl-D-glucose, 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6GDV
 
 | | Structure of CutA from Synechococcus elongatus PCC7942 complexed with Bis-Tris molecule | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
6GDX
 
 | |
6G2W
 
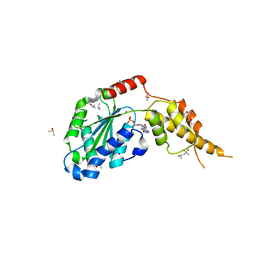 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.678 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G2V
 
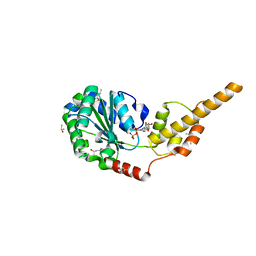 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G4B
 
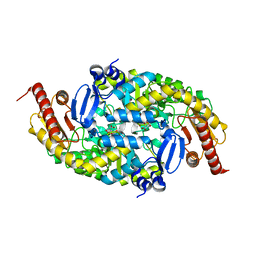 | |
6G2Y
 
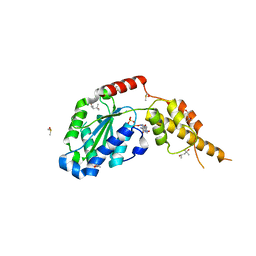 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G4D
 
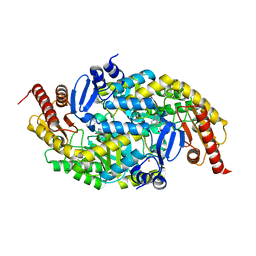 | |
8CHH
 
 | |
8CHG
 
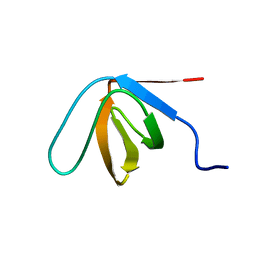 | |
6G2Z
 
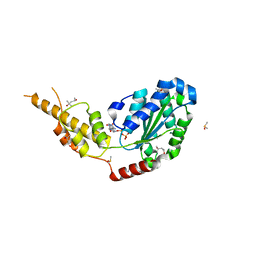 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G4E
 
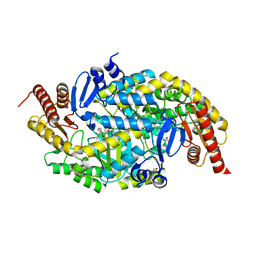 | |
6GDU
 
 | | Structure of CutA from Synechococcus elongatus PCC7942 | | Descriptor: | Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
5LUD
 
 | | Structure of Cyclophilin A in complex with 2,3-Diaminopyridine | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, pyridine-2,3-diamine | | Authors: | McNae, I.W, Nowicki, M.W, Blackburn, E.A, Wear, M.A, Walkinshaw, M.D. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermo-kinetic analysis space expansion for cyclophilin-ligand interactions - identification of a new nonpeptide inhibitor using BiacoreTM T200.
FEBS Open Bio, 7, 2017
|
|
6GG1
 
 | | Structure of PROSS-edited human interleukin 24 | | Descriptor: | Interleukin-24, NICKEL (II) ION, SULFATE ION | | Authors: | Kolenko, P, Zahradnik, J, Kolarova, L, Schneider, B. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Flexible regions govern promiscuous binding of IL-24 to receptors IL-20R1 and IL-22R1.
Febs J., 286, 2019
|
|
