3VIS
 
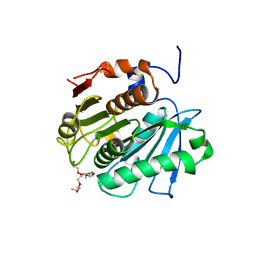 | | Crystal structure of cutinase Est119 from Thermobifida alba AHK119 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Esterase | | Authors: | Kitadokoro, K, Thumarat, U, Nakamura, R, Nishimura, K, Karatani, H, Suzuki, H, Kawai, F. | | Deposit date: | 2011-10-11 | | Release date: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of cutinase Est119 from Thermobida alba AHK119 that can degrade modpolyethylene terephthalate at 1.76 A resolution.
POLYM.DEGRAD.STAB., 97, 2012
|
|
6NQI
 
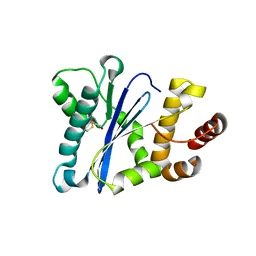 | | Prp8 RH domain from C. merolae | | Descriptor: | BETA-MERCAPTOETHANOL, Pre-mRNA splicing factor PRP8 | | Authors: | Garside, E.L, MacMillan, A.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Prp8 in a Reduced Spliceosome Lacks a Conserved Toggle that Correlates with Splicing Complexity across Diverse Taxa.
J.Mol.Biol., 431, 2019
|
|
6HTJ
 
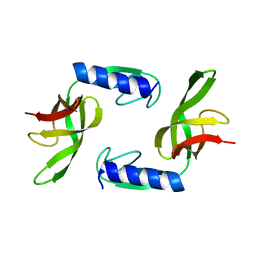 | |
6OKX
 
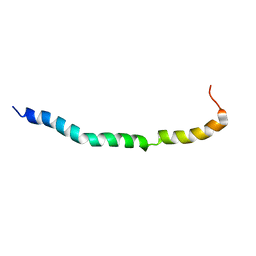 | | Solution structure of VEK50RH1/AA | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
6OKW
 
 | | Solution structure of VEK50 | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
6OQ9
 
 | |
6OKY
 
 | | Solution structure of truncated peptide from PAMap53 | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
3PE6
 
 | |
3PFC
 
 | | Crystal structure of the Lactobacillus johnsonii cinnamoyl esterase LJ0536 S106A mutant in complex with ferulic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Lai, K.K, Vu, C, Xu, X, Cui, H, Molloy, S, Gonzalez, C.F, Yakunin, A, Savchenko, A. | | Deposit date: | 2010-10-28 | | Release date: | 2011-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Inserted alpha/beta Subdomain Shapes the Catalytic Pocket of Lactobacillus johnsonii Cinnamoyl Esterase
Plos One, 6, 2011
|
|
3PF9
 
 | | Crystal structure of the Lactobacillus johnsonii cinnamoyl esterase LJ0536 S106A mutant | | Descriptor: | Cinnamoyl esterase, SODIUM ION | | Authors: | Stogios, P.J, Lai, K.K, Vu, C, Xu, X, Cui, H, Molloy, S, Gonzalez, C.F, Yakunin, A, Savchenko, A. | | Deposit date: | 2010-10-28 | | Release date: | 2011-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Inserted alpha/beta Subdomain Shapes the Catalytic Pocket of Lactobacillus johnsonii Cinnamoyl Esterase
Plos One, 6, 2011
|
|
3PF8
 
 | | Crystal structure of the Lactobacillus johnsonii cinnamoyl esterase LJ0536 | | Descriptor: | Cinnamoyl esterase, SODIUM ION | | Authors: | Stogios, P.J, Lai, K.K, Vu, C, Xu, X, Cui, H, Molloy, S, Gonzalez, C.F, Yakunin, A, Savchenko, A. | | Deposit date: | 2010-10-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | An Inserted alpha/beta Subdomain Shapes the Catalytic Pocket of Lactobacillus johnsonii Cinnamoyl Esterase
Plos One, 6, 2011
|
|
3PFB
 
 | | Crystal structure of the Lactobacillus johnsonii cinnamoyl esterase LJ0536 S106A mutant in complex with ethylferulate | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Cinnamoyl esterase, ... | | Authors: | Stogios, P.J, Lai, K.K, Vu, C, Xu, X, Cui, H, Molloy, S, Gonzalez, C.F, Yakunin, A, Savchenko, A. | | Deposit date: | 2010-10-28 | | Release date: | 2011-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | An Inserted alpha/beta Subdomain Shapes the Catalytic Pocket of Lactobacillus johnsonii Cinnamoyl Esterase
Plos One, 6, 2011
|
|
2MEM
 
 | | Solution NMR structure of SLED domain of Scml2 | | Descriptor: | Sex comb on midleg-like protein 2 | | Authors: | Bezsonova, I. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the DNA-binding Domain from Scml2 (Sex Comb on Midleg-like 2).
J.Biol.Chem., 289, 2014
|
|
2UVP
 
 | | Crystal structure of HobA (HP1230)from Helicobacter pylori | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Terradot, L, Natrajan, G, Thompson, A.C, Hall, D.R. | | Deposit date: | 2007-03-13 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural similarity between the DnaA-binding proteins HobA (HP1230) from Helicobacter pylori and DiaA from Escherichia coli.
Mol. Microbiol., 65, 2007
|
|
6AID
 
 | | Structural insights into the unique polylactate degrading mechanism of Thermobifida alba cutinase | | Descriptor: | CALCIUM ION, Esterase, LACTIC ACID, ... | | Authors: | Kitadokoro, K, Kakara, M, Matsui, S, Osokoshi, R, Thumarat, U, Kawai, F, Kamitani, S. | | Deposit date: | 2018-08-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase.
Febs J., 286, 2019
|
|
2W8M
 
 | |
2MWR
 
 | | Solution Structure of Acidocin B, a Circular Bacteriocin from Lactobacillus acidophilus M46 | | Descriptor: | Acidocin B | | Authors: | Vederas, J.C, Acedo, J.Z, van Belkum, M.J, Lohans, C.T. | | Deposit date: | 2014-11-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Acidocin B, a Circular Bacteriocin Produced by Lactobacillus acidophilus M46.
Appl.Environ.Microbiol., 81, 2015
|
|
6B9M
 
 | |
6AX1
 
 | | Structure of human monoacylglycerol lipase bound to a covalent inhibitor | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-yl 3-(3-phenyl-1,2,4-oxadiazol-5-yl)azetidine-1-carboxylate, GLYCEROL, Monoglyceride lipase | | Authors: | Pandit, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase.
J. Med. Chem., 60, 2017
|
|
7THH
 
 | | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-11 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein
to be published
|
|
2ZZ8
 
 | |
2NA3
 
 | |
3AO9
 
 | | Crystal structure of the C-terminal domain of sequence-specific ribonuclease | | Descriptor: | CADMIUM ION, Colicin-E5 | | Authors: | Inoue, S, Fushinobu, S, Ogawa, T, Hidaka, M, Masaki, H, Yajima, S. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the catalytic residues of sequence-specific and histidine-free ribonuclease colicin E5
J.Biochem., 152, 2012
|
|
3RLI
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 in complex with PMSF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase, phenylmethanesulfonic acid | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
3RM3
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
