7XZG
 
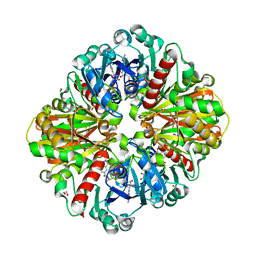 | | High resolution crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans complexed with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cao, H, Meng, J, Ren, Y, Wan, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | High resolution crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans complexed with NAD+
To Be Published
|
|
3RVD
 
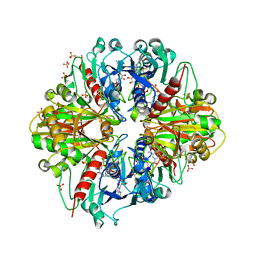 | | Crystal structure of the binary complex, obtained by soaking, of photosyntetic a4 glyceraldehyde 3-phosphate dehydrogenase (gapdh) with cp12-2, both from arabidopsis thaliana. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Thumiger, A, Falini, G, Marri, L, Sparla, F, Trost, P. | | Deposit date: | 2011-05-06 | | Release date: | 2012-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Selection and Folding-upon-binding of Intrinsically Disordered Protein CP12 Regulate Photosynthetic Enzymes Assembly.
J.Biol.Chem., 287, 2012
|
|
3STH
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Toxoplasma gondii | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Staker, B.L, Edwards, T.E, Sankaran, B. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Membrane skeletal association and post-translational allosteric regulation of Toxoplasma gondii GAPDH1.
Mol.Microbiol., 103, 2017
|
|
7Q53
 
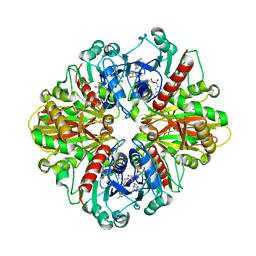 | | Single Particle Cryo-EM structure of photosynthetic A2B2 glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q56
 
 | | Single Particle Cryo-EM structure of photosynthetic A8B8 glyceraldehyde-3-phosphate dehydrogenase (minor conformer) from Spinacia oleracea. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q55
 
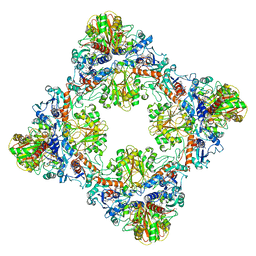 | | Single Particle Cryo-EM structure of photosynthetic A8B8 glyceraldehyde-3-phosphate dehydrogenase hexadecamer (major conformer) from Spinacia oleracia. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q54
 
 | | Single Particle Cryo-EM structure of photosynthetic A4B4-glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q57
 
 | | Single Particle Cryo-EM structure of photosynthetic A10B10 glyceraldehyde-3-phospahte dehydrogenase from Spinacia oleracea. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4LSM
 
 | |
4Z0H
 
 | | X-ray structure of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase (GapC1) complexed with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Orru, R, Falini, G, Trost, P. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tuning Cysteine Reactivity and Sulfenic Acid Stability by Protein Microenvironment in Glyceraldehyde-3-Phosphate Dehydrogenases of Arabidopsis thaliana.
Antioxid. Redox Signal., 24, 2016
|
|
3V1Y
 
 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
4O59
 
 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-19 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4O63
 
 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
3VAZ
 
 | | Crystal structure of Staphylococcal GAPDH1 in a hexagonal space group | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Roychowdhury, A, Mukherjee, S, Dutta, D, Das, A.K. | | Deposit date: | 2011-12-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of Staphylococcal GAPDH1 in a hexagonal space group
To be Published
|
|
4P8R
 
 | |
5C7I
 
 | |
5C7O
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase holo form with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
5C7L
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase apo form | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
3ZDF
 
 | |
3ZCX
 
 | |
5DDI
 
 | |
4QX6
 
 | | CRYSTAL STRUCTURE OF GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM STREPTOCOCCUS AGALACTIAE NEM316 at 2.46 ANGSTROM RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ayres, C.A, Schormann, N, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-07-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase holoenzyme reveals a novel surface.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
1U8F
 
 | |
1CER
 
 | |
1CRW
 
 | |
