1VTO
 
 | |
5EK3
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-[(5~{S})-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
4UB6
 
 | | Native structure of photosystem II (dataset-1) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
9NNO
 
 | | Crystal structure of CYP46A1 with N-[6-(1,3-oxazol-5-yl)-4-(trifluoromethyl)pyridin-2-yl]cyclopropanecarboxamide (compound 4l) | | Descriptor: | 1,2-ETHANEDIOL, Cholesterol 24-hydroxylase, N-[6-(1,3-oxazol-5-yl)-4-(trifluoromethyl)pyridin-2-yl]cyclopropanecarboxamide, ... | | Authors: | Yano, J, Skene, R.J. | | Deposit date: | 2025-03-05 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and identification of brain-penetrant, potent, and selective 1,3-oxazole-based cholesterol 24-hydroxylase (CH24H) inhibitors.
Bioorg.Med.Chem., 124, 2025
|
|
9NNJ
 
 | | Crystal structure of CYP46A1 with 3-chloro-N-[(3M)-3-(1,3-oxazol-5-yl)phenyl]-N-(propan-2-yl)benzene-1-sulfonamide (compound 2) | | Descriptor: | 1,2-ETHANEDIOL, 3-chloro-N-[(3M)-3-(1,3-oxazol-5-yl)phenyl]-N-(propan-2-yl)benzene-1-sulfonamide, Cholesterol 24-hydroxylase, ... | | Authors: | Yano, J, Skene, R.J. | | Deposit date: | 2025-03-05 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Design and identification of brain-penetrant, potent, and selective 1,3-oxazole-based cholesterol 24-hydroxylase (CH24H) inhibitors.
Bioorg.Med.Chem., 124, 2025
|
|
7NNX
 
 | | E. coli NfsA with 1,4-benzoquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, CHLORIDE ION, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
9NNI
 
 | | Crystal structure of CYP46A1 with cyclopropyl[(4M)-4-(1,3-oxazol-5-yl)-6-(trifluoromethyl)-1H-indol-1-yl]methanone (compound 2b) | | Descriptor: | 1,2-ETHANEDIOL, Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yano, J, Skene, R.J. | | Deposit date: | 2025-03-05 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a novel series of cholesterol 24-hydroxylase (CH24H) inhibitors bearing 1,3-oxazole as a heme-iron binding group.
Bioorg.Med.Chem., 128, 2025
|
|
3NKT
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans Adducts with naphthoate | | Descriptor: | 1-hydroxynaphthalene-2-carboxylic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: A step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases.
J.Struct.Biol., 177, 2012
|
|
5EK4
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-[(5~{S})-6-fluoranyl-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, S.Y, Peng, Y.H, Wu, J.S. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
5EK2
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | 1-cyclohexyl-2-[(5~{S})-6-fluoranyl-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanone, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
5ETW
 
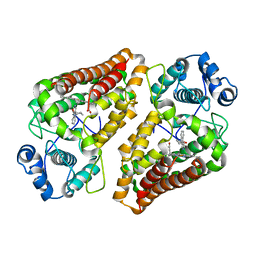 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-pyrido[3,4-b]indol-9-yl-ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, S.Y, Peng, Y.H, Wu, J.S. | | Deposit date: | 2015-11-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1.
J.Med.Chem., 59, 2016
|
|
5V18
 
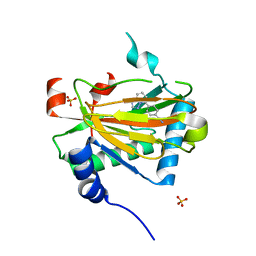 | | Structure of PHD2 in complex with 1,2,4-Triazolo-[1,5-a]pyridine | | Descriptor: | 4-([1,2,4]triazolo[1,5-a]pyridin-5-yl)benzonitrile, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 1,2,4-Triazolo-[1,5-a]pyridine HIF Prolylhydroxylase Domain-1 (PHD-1) Inhibitors With a Novel Monodentate Binding Interaction.
J. Med. Chem., 60, 2017
|
|
5V1B
 
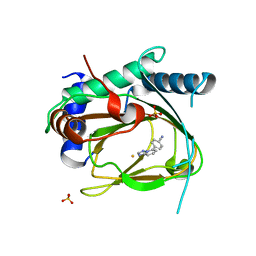 | | Structure of PHD1 in complex with 1,2,4-Triazolo-[1,5-a]pyridine | | Descriptor: | 4-([1,2,4]triazolo[1,5-a]pyridin-5-yl)benzonitrile, Egl nine homolog 2, FE (III) ION, ... | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | 1,2,4-Triazolo-[1,5-a]pyridine HIF Prolylhydroxylase Domain-1 (PHD-1) Inhibitors With a Novel Monodentate Binding Interaction.
J. Med. Chem., 60, 2017
|
|
2WSD
 
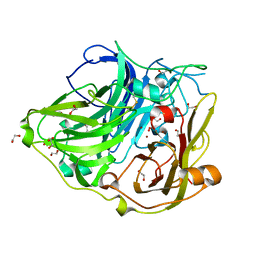 | | Proximal mutations at the type 1 Cu site of CotA-laccase: I494A mutant | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Silva, C.S, Durao, P, Chen, Z, Soares, C.M, Pereira, M.M, Todorovic, S, Hildebrandt, P, Martins, L.O, Lindley, P.F, Bento, I. | | Deposit date: | 2009-09-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proximal Mutations at the Type 1 Copper Site of Cota Laccase: Spectroscopic, Redox, Kinetic and Structural Characterization of I494A and L386A Mutants.
Biochem.J., 412, 2008
|
|
4IL6
 
 | | Structure of Sr-substituted photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Koua, F.H.M, Umena, Y, Kawakami, K, Kamiya, N, Shen, J.R. | | Deposit date: | 2012-12-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Sr-substituted photosystem II at 2.1 A resolution and its implications in the mechanism of water oxidation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3CZY
 
 | |
4HJL
 
 | | Naphthalene 1,2-Dioxygenase bound to 1-chloronaphthalene | | Descriptor: | 1,2-ETHANEDIOL, 1-chloronaphthalene, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-12 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM5
 
 | | Naphthalene 1,2-Dioxygenase bound to indene | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1H-indene, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM4
 
 | | Naphthalene 1,2-Dioxygenase bound to indan | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1H-indene, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM6
 
 | | Naphthalene 1,2-Dioxygenase bound to phenetole | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
1KCW
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN CERULOPLASMIN AT 3.0 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CERULOPLASMIN, COPPER (II) ION, ... | | Authors: | Card, G.L, Zaitsev, V.N, Lindley, P.F. | | Deposit date: | 1996-09-25 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray structure of human serum ceruloplasmin at 3.1 angstrom: Nature of the copper centres.
J.Biol.Inorg.Chem., 1, 1996
|
|
4HKV
 
 | | Naphthalene 1,2-Dioxygenase bound to benzamide | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDE, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM0
 
 | | Naphthalene 1,2-Dioxygenase bound to indole-3-acetate | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
2ORR
 
 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (Delta 114) 4-(Benzo[1,3]dioxol-5-yloxy)-2-(4-imidazol-1-yl-phenoxy)-pyrimidine Complex | | Descriptor: | 1,2-ETHANEDIOL, 4-(1,3-BENZODIOXOL-5-YLOXY)-2-[4-(1H-IMIDAZOL-1-YL)PHENOXY]PYRIMIDINE, Nitric oxide synthase, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
2ORQ
 
 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (DELTA 114) 4-(imidazol-1-yl)phenol and piperonylamine Complex | | Descriptor: | 1-(1,3-BENZODIOXOL-5-YL)METHANAMINE, 4-(1H-IMIDAZOL-1-YL)PHENOL, Nitric oxide synthase, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
