1M2G
 
 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2W
 
 | | Pseudomonas fluorescens mannitol 2-dehydrogenase ternary complex with NAD and D-mannitol | | Descriptor: | D-MANNITOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, mannitol dehydrogenase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2002-06-25 | | Release date: | 2002-11-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pseudomonas fluorescens Mannitol 2-Dehydrogenase Binary and Ternary Complexes. Specificity and Catalytic Mechanism
J.Biol.Chem., 277, 2002
|
|
9GCY
 
 | |
1M3B
 
 | | Solution structure of a circular form of the N-terminal SH3 domain (A134C, E135G, R191G mutant) from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
9GAR
 
 | |
1M3Q
 
 | |
9GSY
 
 | | Cryo-EM structure of mouse PMCA captured in E2-P state (BEF3) | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Vinayagam, D, Raunser, S, Sistel, O, Schulte, U, Constantin, C.E, Prumbaum, D, Zolles, G, Fakler, B. | | Deposit date: | 2024-09-16 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-15 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Molecular mechanism of ultrafast transport by plasma membrane Ca 2+ -ATPases.
Nature, 646, 2025
|
|
1MDZ
 
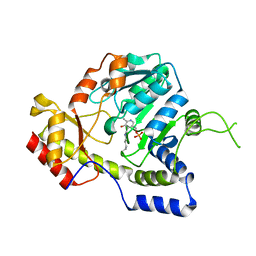 | | Crystal structure of ArnB aminotransferase with cycloserine and pyridoxal 5' phosphate | | Descriptor: | ArnB aminotransferase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
9GD5
 
 | |
9GBU
 
 | | Desulfovibrio desulfuricans [FeFe]-hydrogenase variant with both subunits linked by a 13 amino acid linker peptide derived from a group A1 type [FeFe]-hydrogenase of Solobacterium moorei | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IRON/SULFUR CLUSTER, Periplasmic [Fe] hydrogenase large subunit,Periplasmic [Fe] hydrogenase small subunit, ... | | Authors: | Bikbaev, K, Jaenecke, J, Winkler, M, Span, I. | | Deposit date: | 2024-07-31 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Enhancing maturation of [FeFe]-hydrogenase by interlinking the large and small subunits
To Be Published
|
|
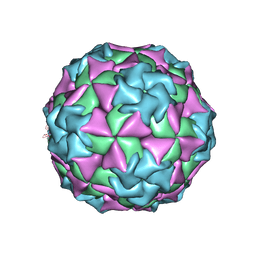
1MEC
 
 | |
1MK0
 
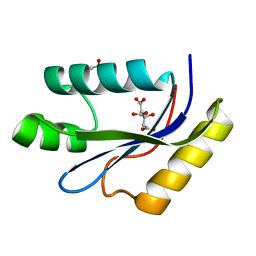 | | catalytic domain of intron endonuclease I-TevI, E75A mutant | | Descriptor: | BETA-MERCAPTOETHANOL, CITRIC ACID, Intron-associated endonuclease 1 | | Authors: | Van Roey, P, Meehan, L, Kowalski, J.C, Belfort, M, Derbyshire, V. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI.
Nat.Struct.Biol., 9, 2002
|
|
9GCP
 
 | | ChREBP/14-3-3 complex stabilized by AMP | | Descriptor: | 14-3-3 protein beta/alpha, N-terminally processed, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Moschref, M, Heimhalt, M, Pysik, T, Menzer, W.M, Basquin, J, Margulies, C.E, Ladurner, A.G. | | Deposit date: | 2024-08-02 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Glucose-6-phosphate functions as a selective receptor agonist for the sugar tolerance transcription factor ChREBP
To Be Published
|
|
9GEU
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,10,11,17,18,21,22-decahydro-4H,16H,20H-13,15-etheno-7,25-(metheno)[1,4]diazepino[2,3-h]pyrido[1,2-p][1]oxa[4,16]diazacyclononadecine-4,8(9H)-dione | | Descriptor: | (R,E)-2-methyl-1,2,3,5,10,11,17,18,21,22-decahydro-4H,16H,20H-13,15-etheno-7,25-(metheno)[1,4]diazepino[2,3-h]pyrido[1,2-p][1]oxa[4,16]diazacyclononadecine-4,8(9H)-dione, (R,R)-2,3-BUTANEDIOL, CITRIC ACID, ... | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the human CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,10,11,17,18,21,22-decahydro-4H,16H,20H-13,15-etheno-7,25-(metheno)[1,4]diazepino[2,3-h]pyrido[1,2-p][1]oxa[4,16]diazacyclononadecine-4,8(9H)-dione
To Be Published
|
|
1MK9
 
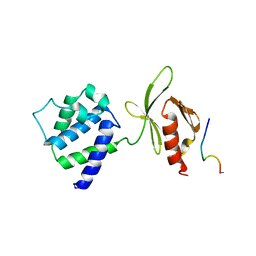 | | CRYSTAL STRUCTURE OF AN INTEGRIN BETA3-TALIN CHIMERA | | Descriptor: | Integrin Beta3, TALIN | | Authors: | Garcia-Alvarez, B, De Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Determinants of Integrin Recognition by Talin
Mol.Cell, 11, 2003
|
|
9GFL
 
 | | Crystal structure of ASO binding Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab fragment heavy chain, ... | | Authors: | Hsia, H.-E, Zanini, C, Simonneau, C, Fraidling, J, Kraft, T, Mayer, K, Sommer, A, Indlekofer, A, Wirth, T, Benz, J, Georges, G, Langer, L.M, Gassner, C, Larraillet, V, Manso, M, Ravn, J, Hofer, K, Emrich, T, Niewoehner, J, Schumacher, F, Brinkmann, U. | | Deposit date: | 2024-08-09 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Improved Targeted Delivery of Antisense Oligonucleotide Conjugates
with the Antibody Mask
To Be Published
|
|
1MF1
 
 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
9GE4
 
 | |
1MFG
 
 | |
7SQE
 
 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with Jun9-84-3 inhibitor | | Descriptor: | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-05 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with Jun9-84-3 inhibitor
To be Published
|
|
1MFL
 
 | |
9GEL
 
 | |
1MFT
 
 | | Crystal Structure Of Four-Helix Bundle Model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Lahr, S.J, Stayrook, S.E, North, B, Kaplan, J, Geremia, S, DeGrado, W. | | Deposit date: | 2002-08-13 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
9GFM
 
 | |
1ML0
 
 | |
