8TWA
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA-PolE-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
7YM1
 
 | | Structure of SsbA protein in complex with the anticancer drug 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Huang, Y.H, Yang, P.C, Chiang, W.Y, Lin, E.S, Huang, C.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil.
Int J Mol Sci, 24, 2023
|
|
7KZ0
 
 | | Human MBD4 glycosylase domain bound to DNA containing substrate analog 2'-deoxy-pseudouridine | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*(P2U)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), GLYCEROL, ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2020-12-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
7EQG
 
 | | Structure of Csy-AcrIF5 | | Descriptor: | AcrIF5, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Nat.Chem.Biol., 18, 2022
|
|
7K5C
 
 | |
4LVK
 
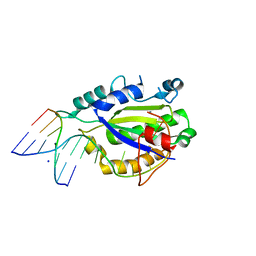 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt+3'Phosphate). Mn-bound crystal structure at pH 4.6 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTGpo oligonucleotide, MANGANESE (II) ION, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8FAY
 
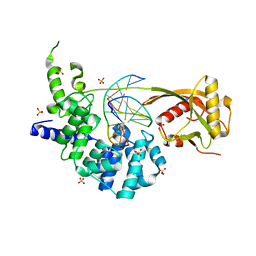 | | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*CP*AP*(NR1)P*GP*TP*CP*T)-3'), ... | | Authors: | Trasvina-Arenas, C.H, Lin, W.J, Demir, M, Fisher, A.J, David, S.S, Horvath, M.P. | | Deposit date: | 2022-11-29 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G)
To Be Published
|
|
1P3I
 
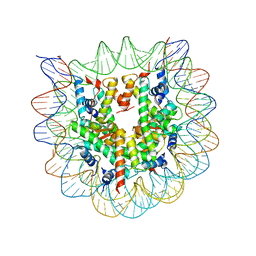 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3M
 
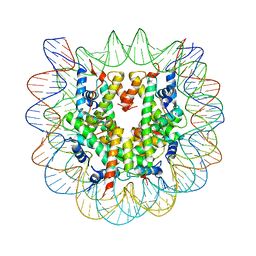 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
2K4L
 
 | | Solution structure of a 2:1C2-(2-naphthyl)pyrrolo[2,1-c][1,4]benzodiazepine (PBD) DNA adduct: molecular basis for unexpectedly high DNA helix stabilization. | | Descriptor: | (11aS)-7,8-dimethoxy-2-naphthalen-2-yl-1,10,11,11a-tetrahydro-5H-pyrrolo[2,1-c][1,4]benzodiazepin-5-one, 5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DAP*DAP*DAP*DGP*DAP*DTP*DT)-3' | | Authors: | Antonow, D, Barata, T, Jenkins, T.C, Parkinson, G.N, Howard, P.W, Thurston, D.E, Zloh, M. | | Deposit date: | 2008-06-13 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a 2:1 C2-(2-naphthyl) pyrrolo[2,1-c][1,4]benzodiazepine DNA adduct: molecular basis for unexpectedly high DNA helix stabilization.
Biochemistry, 47, 2008
|
|
1L2C
 
 | | MutM (Fpg)-DNA Estranged Thymine Mismatch Recognition Complex | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*TP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
6BLW
 
 | |
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
3SBJ
 
 | | MutM slanted complex 7 | | Descriptor: | 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*AP*GP*G)-3', 5'-D(P*CP*CP*TP*GP*GP*TP*(CX)P*TP*AP*CP*C)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Verdine, G.L. | | Deposit date: | 2011-06-05 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Strandwise translocation of a DNA glycosylase on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1P3P
 
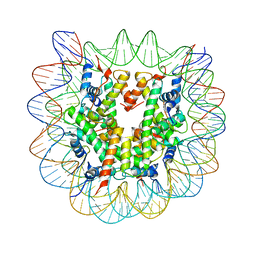 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3G
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
8K28
 
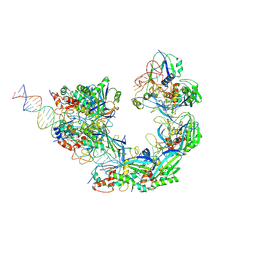 | |
8K29
 
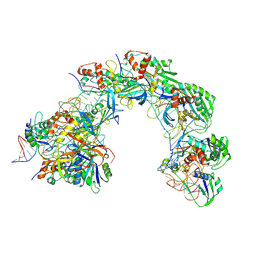 | |
8K27
 
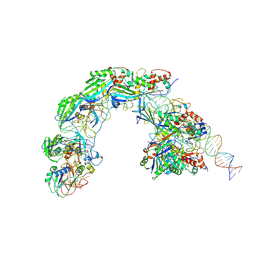 | |
1P34
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
4HRI
 
 | | Crystal structure of HetR in complex with a 21-bp palindromic DNA at the upstream of the hetP promoter from Anabaena | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*GP*AP*CP*CP*CP*CP*TP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*GP*GP*GP*GP*TP*CP*TP*AP*AP*CP*CP*CP*CP*TP*CP*AP*T)-3'), ... | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Chen, Y, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2012-10-28 | | Release date: | 2013-04-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural and biochemical analyses of Anabaena HetR reveal insights into its binding to DNA targets and the inhibitory hexapeptide ERGSGR
To be Published
|
|
1P3B
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3F
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1PAA
 
 | | STRUCTURE OF A HISTIDINE-X4-HISTIDINE ZINC FINGER DOMAIN: INSIGHTS INTO ADR1-UAS1 PROTEIN-DNA RECOGNITION | | Descriptor: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | Authors: | Bernstein, B.E, Hoffman, R.C, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | Deposit date: | 1994-07-15 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a histidine-X4-histidine zinc finger domain: insights into ADR1-UAS1 protein-DNA recognition.
Biochemistry, 33, 1994
|
|
