8VHD
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Isocitrate | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHC
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHB
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Alpha-Ketoglutarate | | Descriptor: | (3~{S})-3-[(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4~{H}-pyridin-4-yl]-2-oxidanylidene-pentanedioic acid, 2-OXOGLUTARIC ACID, CALCIUM ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHA
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Alpha-Ketoglutarate | | Descriptor: | (3~{S})-3-[(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4~{H}-pyridin-4-yl]-2-oxidanylidene-pentanedioic acid, 2-OXOGLUTARIC ACID, CALCIUM ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VH9
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH | | Descriptor: | CITRIC ACID, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VG8
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) Bound to 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG7
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG5
 
 | | Crystal Structure of V113N Variant of D-Dopachrome Tautomerase (D-DT) Bound with 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFW
 
 | | Crystal Structure of V113N D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFU
 
 | |
8VFO
 
 | | Crystal Structure of L117G Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFN
 
 | | Crystal Structure of WT D-Dopachrome Tautomerase (D-DT) at 310K | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFL
 
 | | Crystal Structure of WT D-Dopachrome Tautomerase (D-DT) at 290K | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFK
 
 | | Crystal Structure of Delta 109-117 D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRATE ANION, D-dopachrome decarboxylase, SODIUM ION | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VF6
 
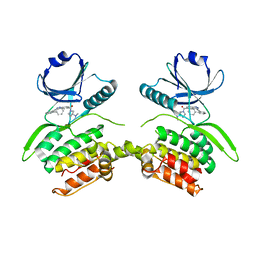 | | Crystal structure of Serine/threonine-protein kinase 33 (STK33) Kinase Domain in complex with inhibitor CDD-2211 | | Descriptor: | Serine/threonine-protein kinase 33, {3-[([1,1'-biphenyl]-2-yl)ethynyl]-1H-indazol-5-yl}[(3R)-3-(dimethylamino)pyrrolidin-1-yl]methanone | | Authors: | Ta, H.M, Kim, C, Ku, K.A, Matzuk, M.M. | | Deposit date: | 2023-12-21 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reversible male contraception by targeted inhibition of serine/threonine kinase 33.
Science, 384, 2024
|
|
8VEX
 
 | |
8VEW
 
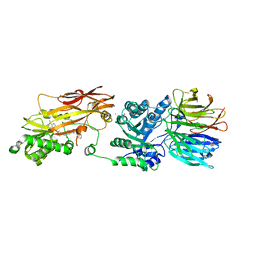 | | Crystal structure of PRMT5:MEP50 in complex with MTA and oxamide compound 24 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-{2-[(2R,5S)-5-methyl-2-phenylpiperidin-1-yl](oxo)acetamido}pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEO
 
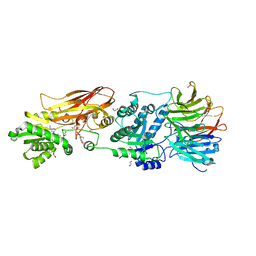 | | Crystal structure of PRMT5:MEP50 in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEK
 
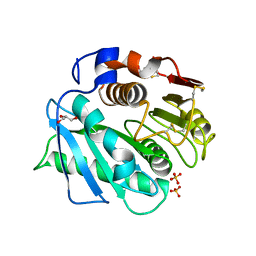 | | IsPETase - ACC mutant | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
8VEH
 
 | |
8VE9
 
 | | IsPETase - ACCCETN mutant - CombiPETase | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-18 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
8VDY
 
 | | Crystal Structure of Delta 114-117 D-Dopachrome Tautomerase (D-DT) | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VDB
 
 | |
8VD9
 
 | |
8VCW
 
 | | X-Ray Crystal Structure of the biotin synthase from B. obeum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, Biotin synthase, ... | | Authors: | Lachowicz, J.C, Grove, T.L. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Biotin Synthase That Utilizes an Auxiliary 4Fe-5S Cluster for Sulfur Insertion.
J.Am.Chem.Soc., 146, 2024
|
|
