9FZX
 
 | | Rhizobium phage ligase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, DNA, ... | | Authors: | Rothweiler, U, Williamson, A. | | Deposit date: | 2024-07-06 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Crystal structure of ATP-dependent DNA ligase from Rhizobium phage vB_RleM_P10VF.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
1M2R
 
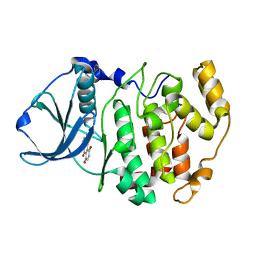 | | Crystal structure of 5,8-di-amino-1,4-di-hydroxy-anthraquinone/CK2 kinase complex | | Descriptor: | 5,8-DI-AMINO-1,4-DIHYDROXY-ANTHRAQUINONE, CASEIN kinase II, alpha chain | | Authors: | De Moliner, E, Moro, S, Sarno, S, Zagotto, G, Zanotti, G, Pinna, L.A, Battistutta, R. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of protein kinase CK2 by anthraquinone-related compounds. A
structural insight
J.Biol.Chem., 278, 2003
|
|
9GD9
 
 | |
1LVR
 
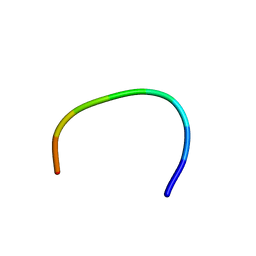 | | IC3 of CB1 (L431A,A432L) Bound to G(alpha)i | | Descriptor: | Cannabinoid receptor 1 | | Authors: | Ulfers, A.L, McMurry, J.L, Miller, A, Wang, L, Kendall, D.A, Mierke, D.F. | | Deposit date: | 2002-05-29 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Cannabinoid receptor-G protein interactions: G(alphai1)-bound structures of IC3 and a mutant with altered G protein specificity.
Protein Sci., 11, 2002
|
|
1M34
 
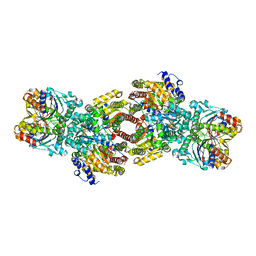 | | Nitrogenase Complex From Azotobacter Vinelandii Stabilized By ADP-Tetrafluoroaluminate | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schmid, B, Einsle, O, Chiu, H.-J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-27 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4 Stabilized structure
Biochemistry, 41, 2002
|
|
7RX8
 
 | |
1M5O
 
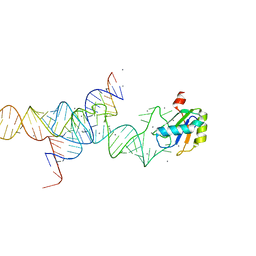 | | Transition State Stabilization by a Catalytic RNA | | Descriptor: | CALCIUM ION, RNA SUBSTRATE, RNA HAIRPIN RIBOZYME, ... | | Authors: | Rupert, P.B, Massey, A.P, Sigurdsson, S.T, Ferre-D'Amare, A.R. | | Deposit date: | 2002-07-09 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transition state stabilization by a catalytic RNA
Science, 298, 2002
|
|
9GND
 
 | |
9G88
 
 | | Carotenoid cleavage oxygenase from Moesziomyces aphidis bound to acetate | | Descriptor: | ACETATE ION, FE (II) ION, Lignostilbene dioxygenase | | Authors: | Plewka, J, Schober, L, Magiera-Mularz, K, Rudroff, F, Winkler, M. | | Deposit date: | 2024-07-23 | | Release date: | 2025-06-25 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characteristics of Potent Dioxygenase from Moesziomyces aphidis .
Jacs Au, 5, 2025
|
|
1LWS
 
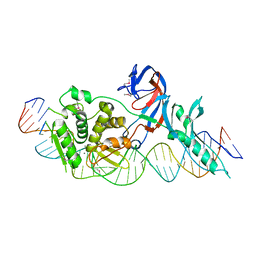 | | Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence | | Descriptor: | CALCIUM ION, ENDONUCLEASE PI-SCEI, PI-SceI DNA recognition region bottom strand, ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence.
Nat.Struct.Biol., 9, 2002
|
|
1LXE
 
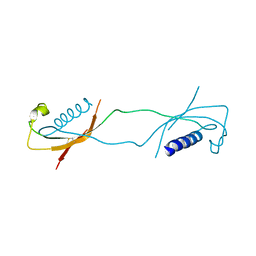 | | CRYSTAL STRUCTURE OF THE CATHELICIDIN MOTIF OF PROTEGRINS | | Descriptor: | protegrin-3 precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-06-05 | | Release date: | 2002-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
1M61
 
 | |
9GMS
 
 | | Mtb PNPase Rv2783c | | Descriptor: | 1-[[4-[4-[[2-phenyl-5-(trifluoromethyl)-1,3-oxazol-4-yl]carbonylamino]phenyl]phenyl]carbonylamino]cyclopentane-1-carboxylic acid, Polyribonucleotide nucleotidyltransferase, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Griesser, T, Sander, P. | | Deposit date: | 2024-08-29 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Selective inhibition of Mycobacterium tuberculosis GpsI unveils a novel strategy to target the RNA metabolism.
Nucleic Acids Res., 53, 2025
|
|
9GTK
 
 | | KRAS in complex with DARPin 784_F5 | | Descriptor: | 1,2-ETHANEDIOL, DARPin 784_F5, Isoform 2B of GTPase KRas, ... | | Authors: | Kapp, J.N, Verdurmen, W, Schaefer, J.V, Kopra, K, Nagy-Davidescu, G, Richard, E, Nokin, M.J, Ernst, P, Tamaskovic, R, Schwill, M, Degen, R, Scholl, C, Santamaria, D, Plueckthun, A. | | Deposit date: | 2024-09-18 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A nucleotide-independent, pan-RAS-targeted DARPin elicits anti-tumor activity in a multimodal manner.
Mol Oncol, 2025
|
|
1M66
 
 | | Crystal Structure of Leishmania mexicana GPDH Complexed with Inhibitor 2-bromo-6-chloro-purine | | Descriptor: | 2-BROMO-6-CHLORO-PURINE, Glycerol-3-phosphate dehydrogenase, PALMITIC ACID | | Authors: | Choe, J, Suresh, S, Wisedchaisri, G, Kennedy, K.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2002-07-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anomalous differences of light elements in determining precise binding modes of ligands to
glycerol-3-phosphate dehydrogenase
Chem.Biol., 9, 2002
|
|
9GMT
 
 | | Mtb PNPase Rv2783c Mutant L328F | | Descriptor: | 1-[[4-[4-[[2-phenyl-5-(trifluoromethyl)-1,3-oxazol-4-yl]carbonylamino]phenyl]phenyl]carbonylamino]cyclopentane-1-carboxylic acid, Polyribonucleotide nucleotidyltransferase, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Griesser, T, Sander, P. | | Deposit date: | 2024-08-29 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Selective inhibition of Mycobacterium tuberculosis GpsI unveils a novel strategy to target the RNA metabolism.
Nucleic Acids Res., 53, 2025
|
|
1M6H
 
 | | Human glutathione-dependent formaldehyde dehydrogenase | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-16 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
1LY4
 
 | | Analysis of quinazoline and PYRIDO[2,3D]PYRIMIDINE N9-C10 reversed bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
9GM0
 
 | |
7RYW
 
 | | Crystal structure of Ferritin grown by the microbatch method in the presence of Agarose | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, D.J, Stojanoff, V. | | Deposit date: | 2021-08-26 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of Ferritin grown by microbatch method in the presence of agarose
To Be Published
|
|
9G6T
 
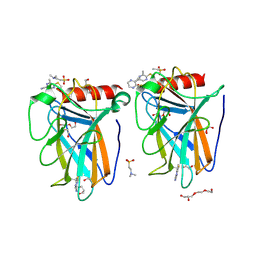 | | p53-Y220C Core Domain Covalently Bound to 5-Chloro-6-methylpyrazine-2-carbonitrile Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chloranyl-6-methyl-pyrazine-2-carbonitrile, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | S N Ar Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes.
Drug Des Devel Ther, 19, 2025
|
|
1LYZ
 
 | |
1M76
 
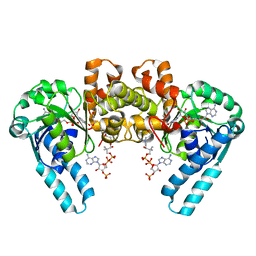 | |
9GC8
 
 | | Crystal structure of Pseudomonas aeruginosa IspD in complex with C11H12N2O3 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, methyl 2-[(5~{R})-6-methyl-7-oxidanylidene-5~{H}-pyrrolo[3,4-b]pyridin-5-yl]ethanoate | | Authors: | Borel, F, D'Auria, L. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment Discovery by X-Ray Crystallographic Screening Targeting the CTP Binding Site of Pseudomonas Aeruginosa IspD.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
1M7B
 
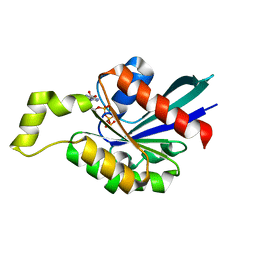 | | Crystal structure of Rnd3/RhoE: functional implications | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rnd3/RhoE small GTP-binding protein | | Authors: | Fiegen, D, Blumenstein, L, Stege, P, Vetter, I.R, Ahmadian, M.R. | | Deposit date: | 2002-07-19 | | Release date: | 2002-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Rnd3/RhoE: functional implications
FEBS LETT., 525, 2002
|
|
