5WSQ
 
 | | Crystal structure of C-Hg-T pair containing DNA duplex | | Descriptor: | 1,3-PROPANDIOL, DNA (5'-D(*GP*CP*CP*CP*GP*TP*GP*C)-3'), MERCURY (II) ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
2Z2W
 
 | | Human Wee1 kinase complexed with inhibitor PF0335770 | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[4-(2-CHLOROPHENYL)-1,3-DIOXO-1,2,3,6-TETRAHYDROPYRROLO[3,4-C]CARBAZOL-9-YL]FORMAMIDE, ... | | Authors: | Squire, C.J, Baker, E.N. | | Deposit date: | 2007-05-29 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Synthesis and Structure-Activity Relationships of 9-Amino-4-(2-chlorophenyl)pyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones and Related Formamides as Inhibitors of the Wee1 and Chk1 Checkpoint Kinases
To be Published
|
|
6SUV
 
 | |
8UMC
 
 | | Deinococcus aerius TR0125 C-glucosyl deglycosidase (CGD), cryo-EM | | Descriptor: | DUF6379 domain-containing protein, MANGANESE (II) ION, Xylose isomerase-like TIM barrel domain-containing protein | | Authors: | Furlanetto, V, Kalyani, D.C, Kostelac, A, Haltrich, D, Hallberg, B.M, Divne, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius.
J.Mol.Biol., 436, 2024
|
|
2XYM
 
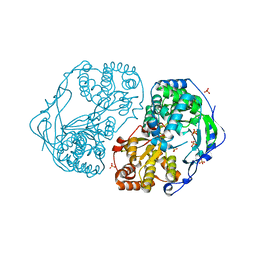 | | HCV-JFH1 NS5B T385A mutant | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Simister, P.C, Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
1XOY
 
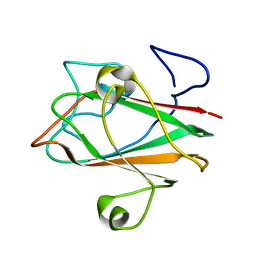 | | Solution structure of At3g04780.1, an Arabidopsis ortholog of the C-terminal domain of human thioredoxin-like protein | | Descriptor: | hypothetical protein At3g04780.1 | | Authors: | Song, J, Robert, C.T, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of At3g04780.1-des15, an Arabidopsis thaliana ortholog of the C-terminal domain of human thioredoxin-like protein.
Protein Sci., 14, 2005
|
|
3ZJ0
 
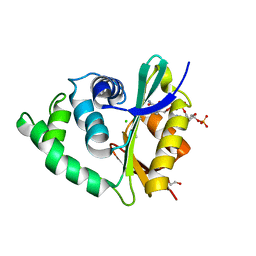 | | The human O-GlcNAcase C-terminal domain is a pseudo histone acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, ACETYLTRANSFERASE, ... | | Authors: | Rao, F.V, Schuettelkopf, A.W, Dorfmueller, H.C, Ferenbach, A.T, Navratilova, I, van Aalten, D.M.F. | | Deposit date: | 2013-01-15 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Bacterial Putative Acetyltransferase Defines the Fold of the Human O-Glcnacase C-Terminal Domain.
Open Biol., 3, 2013
|
|
2LA4
 
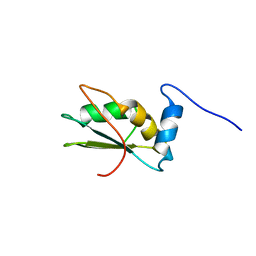 | | NMR structure of the C-terminal RRM domain of poly(U) binding 1 | | Descriptor: | Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Santiveri, C.M, Mirassou, Y, Rico-Lastres, P, Martinez-Lumbreras, S, Perez-Canadillas, J.M. | | Deposit date: | 2011-03-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pub1p C-terminal RRM domain interacts with Tif4631p through a conserved region neighbouring the Pab1p binding site
Plos One, 6, 2011
|
|
3K4G
 
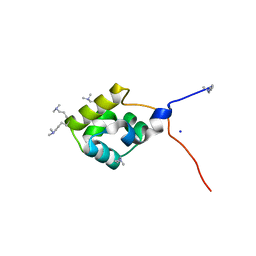 | |
1QAD
 
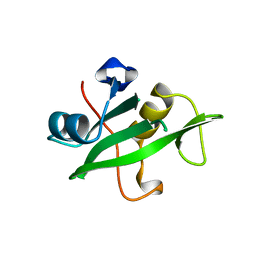 | | Crystal Structure of the C-Terminal SH2 Domain of the P85 alpha Regulatory Subunit of Phosphoinositide 3-Kinase: An SH2 domain mimicking its own substrate | | Descriptor: | PI3-KINASE P85 ALPHA SUBUNIT | | Authors: | Hoedemaeker, P.J, Siegal, G, Roe, M, Driscoll, P.C, Abrahams, J.P.A. | | Deposit date: | 1999-02-26 | | Release date: | 1999-10-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C-terminal SH2 domain of the p85alpha regulatory subunit of phosphoinositide 3-kinase: an SH2 domain mimicking its own substrate.
J.Mol.Biol., 292, 1999
|
|
7UJ6
 
 | | Outer Surface Protein C Type K | | Descriptor: | Outer surface protein C | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2022-03-30 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural Elucidation of a Protective B Cell Epitope on Outer Surface Protein C (OspC) of the Lyme Disease Spirochete, Borreliella burgdorferi.
Mbio, 14, 2023
|
|
1V3X
 
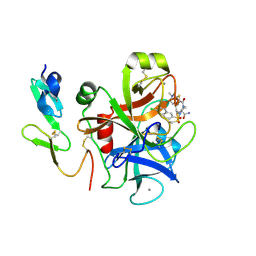 | | Factor Xa in complex with the inhibitor 1-[6-methyl-4,5,6,7-tetrahydrothiazolo(5,4-c)pyridin-2-yl] carbonyl-2-carbamoyl-4-(6-chloronaphth-2-ylsulphonyl)piperazine | | Descriptor: | (2R)-4-[(6-CHLORO-2-NAPHTHYL)SULFONYL]-1-[(5-METHYL-4,5,6,7-TETRAHYDRO[1,3]THIAZOLO[5,4-C]PYRIDIN-2-YL)CARBONYL]PIPERAZ INE-2-CARBOXAMIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and conformational analysis of a non-amidine factor Xa inhibitor that incorporates 5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridine as S4 binding element
J.Med.Chem., 47, 2004
|
|
2A4W
 
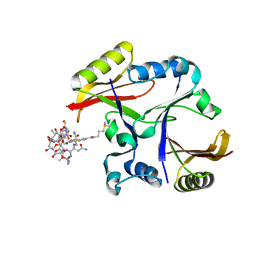 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Copper(II)-Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, COPPER (II) ION, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
2A4X
 
 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Metal-Free Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
1UND
 
 | | Solution structure of the human advillin C-terminal headpiece subdomain | | Descriptor: | ADVILLIN | | Authors: | Vermeulen, W, Van Troys, M, Vanhaesebrouck, P, Verschueren, M, Fant, F, Ampe, C, Martins, J, Borremans, F. | | Deposit date: | 2003-09-09 | | Release date: | 2004-07-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the C-Terminal Headpiece Subdomains of Human Villin and Advillin, Evaluation of Headpiece F-Actin-Binding Requirements
Protein Sci., 13, 2004
|
|
2AX0
 
 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5x) | | Descriptor: | 5R-(2E-METHYL-3-PHENYL-ALLYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
2QZ6
 
 | | First crystal structure of a psychrophile class C beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Michaux, C, Massant, J, Kerff, F, Charlier, P, Wouters, J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of a cold-adapted class C beta-lactamase
Febs J., 275, 2008
|
|
5AUS
 
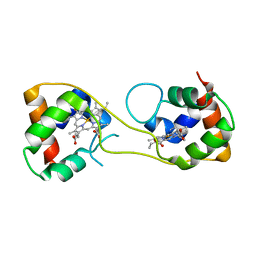 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at C-terminal region | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
1C7M
 
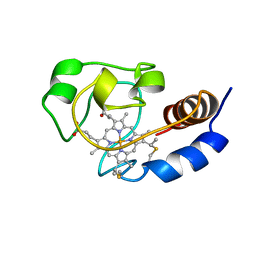 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | HEME C, PROTEIN (CYTOCHROME C552) | | Authors: | Pristovsek, P, Luecke, C, Reincke, B, Ludwig, B, Rueterjans, H. | | Deposit date: | 2000-03-03 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the functional domain of Paracoccus denitrificans cytochrome c552 in the reduced state.
Eur.J.Biochem., 267, 2000
|
|
2XHU
 
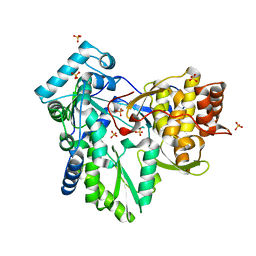 | | HCV-J4 NS5B Polymerase Orthorhombic Crystal Form | | Descriptor: | RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2CE0
 
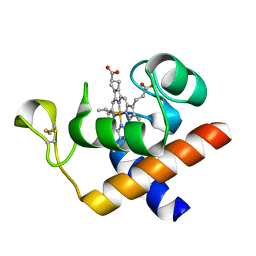 | | Structure of oxidized Arabidopsis thaliana cytochrome 6A | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Marcaida, M.J, Schlarb-Ridley, B.G, Worrall, J.A.R, Wastl, J, Evans, T.J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2006-02-01 | | Release date: | 2006-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure of Cytochrome C(6A), a Novel Dithio-Cytochrome of Arabidopsis Thaliana, and its Reactivity with Plastocyanin: Implications for Function.
J.Mol.Biol., 360, 2006
|
|
1AH7
 
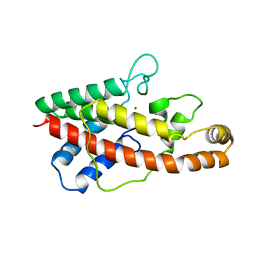 | | PHOSPHOLIPASE C FROM BACILLUS CEREUS | | Descriptor: | PHOSPHOLIPASE C, ZINC ION | | Authors: | Greaves, R. | | Deposit date: | 1997-04-14 | | Release date: | 1997-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | High-resolution (1.5 A) crystal structure of phospholipase C from Bacillus cereus.
Nature, 338, 1989
|
|
2JNF
 
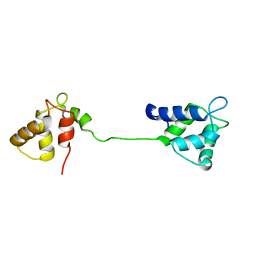 | |
3LLU
 
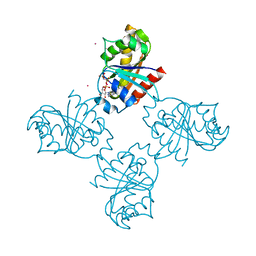 | | Crystal structure of the nucleotide-binding domain of Ras-related GTP-binding protein C | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related GTP-binding protein C, ... | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Crombet, L, Zhong, N, Guan, X, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the nucleotide-binding domain of Ras-related GTP-binding protein C
to be published
|
|
5EXX
 
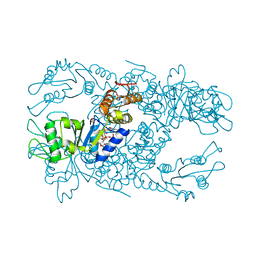 | | AAA+ ATPase FleQ from Pseudomonas aeruginosa bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Transcriptional regulator FleQ | | Authors: | Navarro, M.V.A.S, Sondermann, H, Krasteva, P.V. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.311 Å) | | Cite: | Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
