7MSC
 
 | | Mtb 70SIC in complex with MtbEttA at Pre_R0 state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MT7
 
 | | Mtb 70S with P and E site tRNAs | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-13 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MT3
 
 | | Mtb 70S with P/E tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MPJ
 
 | | Stm1 bound vacant 80S structure isolated from wild-type | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
9O3L
 
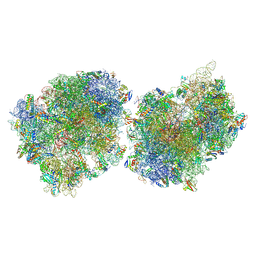 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolide erythromycin, mRNA, deacylated A-site tRNAphe, P-site fMRC-peptidyl-tRNAmet, and deacylated E-site tRNAphe at 2.75A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Kruglov, A.A, Paranjpe, M.N, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2025-04-07 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into context-specific inhibition of bacterial translation by macrolides.
Biorxiv, 2025
|
|
4V88
 
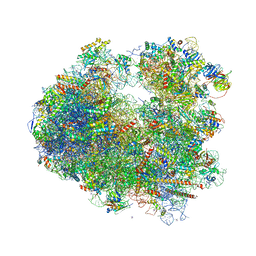 | | The structure of the eukaryotic ribosome at 3.0 A resolution. | | Descriptor: | 18S RIBOSOMAL RNA, 18S rRNA, 25S rRNA, ... | | Authors: | Ben-Shem, A, Garreau de Loubresse, N, Melnikov, S, Jenner, L, Yusupova, G, Yusupov, M. | | Deposit date: | 2011-10-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the eukaryotic ribosome at 3.0 angstrom resolution.
Science, 334, 2011
|
|
4V5D
 
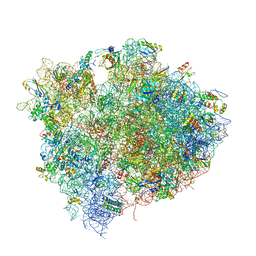 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
4V5T
 
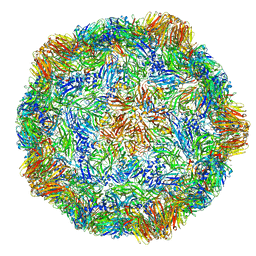 | | X-ray structure of the Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Sauter, C, Lorber, B, Bron, P, Trapani, S, Bergdoll, M, Marmonier, A, Schmitt-Keichinger, C, Lemaire, O, Demangeat, G, Ritzenthaler, C. | | Deposit date: | 2011-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights Into Viral Determinants of Nematode Mediated Grapevine Fanleaf Virus Transmission.
Plos Pathog., 7, 2011
|
|
9MTR
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site GGS-mutant Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTQ
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site GGD-mutant Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTS
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-unmodified Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9NO7
 
 | | Cryo-EM structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site 2'-deoxy-A76-fMEAAAKC-peptidyl-tRNAcys at 2.13A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-03-08 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTP
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTT
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site deacylated-tRNAcys at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9NYO
 
 | |
9Q87
 
 | | Principles of ion binding to RNA inferred from the analysis of a 1.55 Angstrom resolution bacterial ribosome structure - Part I: Mg2+ | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Leonarski, f, Henning-Knechtel, A, Kirmizialtin, S, Ennifar, E, Auffinger, P. | | Deposit date: | 2025-02-23 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | Principles of ion binding to RNA inferred from the analysis of a 1.55 Angs. resolution bacterial ribosome structure - Part I: Mg2+.
Nucleic Acids Res, 2024
|
|
9O3I
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with ketolide telithromycin, mRNA, aminoacylated A-site Lys-tRNAlys, P-site fMRC-peptidyl-tRNAmet, and deacylated E-site tRNAlys at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Kruglov, A.A, Paranjpe, M.N, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2025-04-07 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into context-specific inhibition of bacterial translation by macrolides.
Biorxiv, 2025
|
|
4V5C
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A-site tRNA, deacylated P-site tRNA, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Substrate Stabilization from Snapshots of the Peptidyl Transferase Center of the Intact 70S Ribosome
Nat.Struct.Mol.Biol., 16, 2009
|
|
9O3K
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolide erythromycin, mRNA, aminoacylated A-site Lys-tRNAlys, P-site fMAC-peptidyl-tRNAmet, and deacylated E-site tRNAlys at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Kruglov, A.A, Paranjpe, M.N, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2025-04-07 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into context-specific inhibition of bacterial translation by macrolides.
Biorxiv, 2025
|
|
9O3J
 
 | | Crystal structure of the wild-type drug-free Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Lys-tRNAlys, P-site fMRC-peptidyl-tRNAmet, and deacylated E-site tRNAlys at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Kruglov, A.A, Paranjpe, M.N, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2025-04-07 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into context-specific inhibition of bacterial translation by macrolides.
Biorxiv, 2025
|
|
9O3H
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolide erythromycin, mRNA, aminoacylated A-site Lys-tRNAlys, P-site fMRC-peptidyl-tRNAmet, and deacylated E-site tRNAlys at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Kruglov, A.A, Paranjpe, M.N, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2025-04-07 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into context-specific inhibition of bacterial translation by macrolides.
Biorxiv, 2025
|
|
9L7E
 
 | | Crystal structure of human kinesin-1 motor domain (G234A mutant) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-1 heavy chain, MAGNESIUM ION | | Authors: | Makino, T, Miyazono, K, Tanokura, M, Tomishige, M. | | Deposit date: | 2024-12-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tension-induced suppression of allosteric conformational changes coordinates kinesin-1 stepping.
J.Cell Biol., 224, 2025
|
|
9R86
 
 | |
9R87
 
 | |
4V9R
 
 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
