3ITY
 
 | | Metal-free form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITZ
 
 | |
3IU0
 
 | |
3IU1
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA
To be Published
|
|
3IU2
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096 | | Descriptor: | (2R)-2-{4-hydroxy-5-methoxy-2-[3-(4-methylpiperazin-1-yl)propyl]phenyl}-3-pyridin-3-yl-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096
To be Published
|
|
3IU3
 
 | | Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of Basiliximab, Interleukin-2 receptor alpha chain, Light chain of Fab fragment of Basiliximab, ... | | Authors: | Du, J, Yang, H, Wang, J, Ding, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab
J.Immunol., 184, 2010
|
|
3IU4
 
 | | anti NeuGcGM3 ganglioside chimeric antibody chP3 | | Descriptor: | SULFATE ION, chP3 Fab heavy chain, chP3 Fab light chain | | Authors: | Eriksson, A, Okvist, M, Talavera, A, Krengel, U. | | Deposit date: | 2009-08-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of an anti-ganglioside antibody, and modelling of the functional mimicry of its NeuGc-GM3 antigen by an anti-idiotypic antibody.
Mol.Immunol., 46, 2009
|
|
3IU5
 
 | | Crystal structure of the first bromodomain of human poly-bromodomain containing protein 1 (PB1) | | Descriptor: | Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-30 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3IU6
 
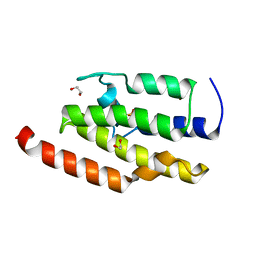 | | Crystal structure of the sixth bromodomain of human poly-bromodomain containing protein 1 (PB1) | | Descriptor: | 1,2-ETHANEDIOL, Protein polybromo-1, ZINC ION | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-30 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3IU7
 
 | |
3IU8
 
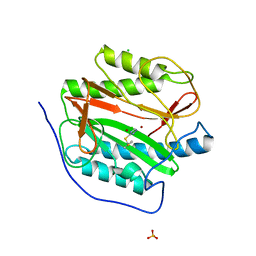 | | M. tuberculosis methionine aminopeptidase with Ni inhibitor T03 | | Descriptor: | 3-[(4-fluorobenzyl)sulfanyl]-4H-1,2,4-triazole, CHLORIDE ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2009-08-30 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalysis and Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidase
J.Med.Chem., 53, 2010
|
|
3IU9
 
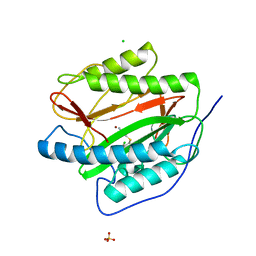 | | M. tuberculosis methionine aminopeptidase with Ni inhibitor T07 | | Descriptor: | 5-[(2,4-dichlorobenzyl)sulfanyl]-4H-1,2,4-triazol-3-amine, CHLORIDE ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2009-08-30 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis and Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidase
J.Med.Chem., 53, 2010
|
|
3IUB
 
 | | Crystal structure of pantothenate synthetase from Mycobacterium tuberculosis in complex with 5-Methoxy-N-(5-methylpyridin-2-ylsulfonyl)-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-methoxy-N-[(5-methylpyridin-2-yl)sulfonyl]-1H-indole-2-carboxamide, ETHANOL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IUC
 
 | | Crystal structure of the human 70kDa heat shock protein 5 (BiP/GRP78) ATPase domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Heat shock 70kDa protein 5 (Glucose-regulated protein, ... | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
3IUD
 
 | | Cu2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COPPER (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUE
 
 | | Crystal structure of pantothenate synthetase in complex with 2-(5-methoxy-2-(5-Methylpyridin-2-ylsulfonylcarbamoyl)-1H-indol-1-yl) acetic acid | | Descriptor: | (5-methoxy-2-{[(5-methylpyridin-2-yl)sulfonyl]carbamoyl}-1H-indol-1-yl)acetic acid, 1,2-ETHANEDIOL, ETHANOL, ... | | Authors: | Silvestre, H.L, Wen, S, Hung, A.W, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IUF
 
 | | Crystal structure of the C2H2-type zinc finger domain of human ubi-d4 | | Descriptor: | ZINC ION, Zinc finger protein ubi-d4 | | Authors: | Tempel, W, Xu, C, Bian, C, Adams-Cioaba, M, Eryilmaz, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Cys2His2-type zinc finger domain of human DPF2.
Biochem.Biophys.Res.Commun., 413, 2011
|
|
3IUG
 
 | | Crystal structure of the RhoGAP domain of RICS | | Descriptor: | Rho/Cdc42/Rac GTPase-activating protein RICS, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the RhoGAP domain of RICS
to be published
|
|
3IUH
 
 | | Co2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COBALT (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUI
 
 | | Zn2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUJ
 
 | | apPEP_WT2 opened state | | Descriptor: | Prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUK
 
 | | Crystal structure of putative bacterial protein of unknown function (DUF885, PF05960.1, ) from Arthrobacter aurescens TC1, reveals fold similar to that of M32 carboxypeptidases | | Descriptor: | GLYCEROL, MAGNESIUM ION, uncharacterized protein | | Authors: | Nocek, B, Chhor, G, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative bacterial protein of unknown function (DUF885, PF05960.1, ) from Arthrobacter aurescens TC1, reveals fold similar to that of M32 carboxypeptidases
To be Published
|
|
3IUL
 
 | | apPEP_WT1 opened state | | Descriptor: | GLYCEROL, Prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUM
 
 | | apPEP_WTX opened state | | Descriptor: | GLYCEROL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUN
 
 | | apPEP_D622N opened state | | Descriptor: | GLYCEROL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
