7S8A
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE, CUBIC CRYSTAL FORM | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S79
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH SYNTHETIC PHOSPHONO-MLL PEPTIDE ANALOG | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S7D
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH SYNTHETIC SULFO-MLL PEPTIDE ANALOG | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8F
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8E
 
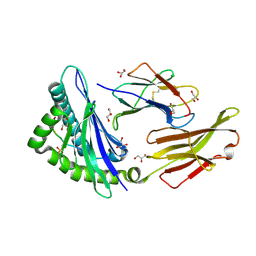 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7Z1Y
 
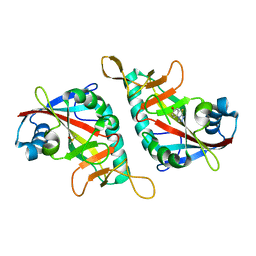 | | PARP15 catalytic domain in complex with OUL245 | | Descriptor: | DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15, [1,2,4]triazolo[3,4-b][1,3]benzothiazol-6-ol | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7Z1V
 
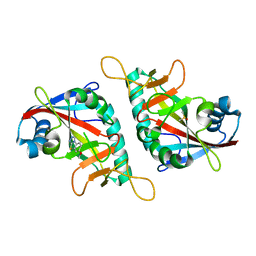 | | PARP15 catalytic domain in complex with OUL208 | | Descriptor: | 6-methoxy-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7Z2O
 
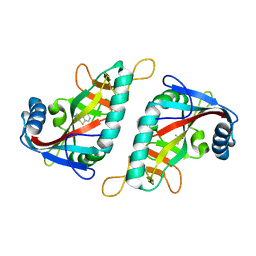 | | PARP15 catalytic domain in complex with OUL215 | | Descriptor: | 8-methoxy-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7Z41
 
 | | PARP15 catalytic domain in complex with OUL209 | | Descriptor: | 5,8-dimethoxy-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Murthy, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7Z2Q
 
 | | PARP15 catalytic domain in complex with OUL232 | | Descriptor: | 5,8-dimethoxy-[1,2,4]triazolo[3,4-b][1,3]benzothiazol-1-amine, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
6MFJ
 
 | |
7Z1W
 
 | | PARP15 catalytic domain in complex with OUL246 | | Descriptor: | 6-chloranyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
3HUQ
 
 | | Thieno[3,2-b]thiophene in complex with T4 lysozyme L99A/M102Q | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
7CF8
 
 | | PfkB(Mycobacterium marinum) | | Descriptor: | Fructokinase, PfkB, GLYCEROL, ... | | Authors: | Li, J, Gao, B, Ji, R. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural analysis and functional study of phosphofructokinase B (PfkB) from Mycobacterium marinum.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
5EIU
 
 | | Mini TRIM5 B-box 2 dimer C2 crystal form | | Descriptor: | TRIM protein-E3 ligase Chimera, ZINC ION | | Authors: | Wagner, J.M, Doss, G, Pornillos, O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5 alpha.
Elife, 5, 2016
|
|
6S2Z
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Brassica oleracea var. Botrytis with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-Soluble Chlorophyll Protein | | Authors: | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | Deposit date: | 2019-06-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
6ORO
 
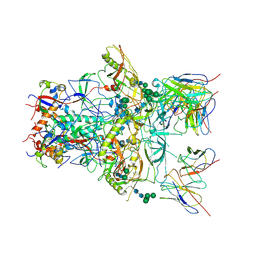 | | Modified BG505 SOSIP-based immunogen RC1 in complex with the elicited V3-glycan patch antibody Ab874NHP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab874NHP antibody Fab heavy chain, ... | | Authors: | Abernathy, M.E, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques.
Nature, 570, 2019
|
|
5OOG
 
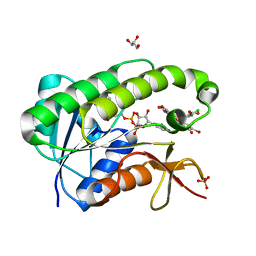 | | Human biliverdin IX beta reductase: NADP/Phloxine B ternary complex | | Descriptor: | Flavin reductase (NADPH), GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | In silicoand crystallographic studies identify key structural features of biliverdin IX beta reductase inhibitors having nanomolar potency.
J. Biol. Chem., 293, 2018
|
|
2JIE
 
 | |
8T5Q
 
 | |
6TH4
 
 | | Tubulin-inhibitor complex | | Descriptor: | 1,2,3,9-tetramethoxy-6-methylidene-5~{H}-cyclohepta[a]naphthalen-8-one, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Varela, P.F, Gigant, B. | | Deposit date: | 2019-11-18 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | B-nor-methylene Colchicinoid PT-100 Selectively Induces Apoptosis in Multidrug-Resistant Human Cancer Cells via an Intrinsic Pathway in a Caspase-Independent Manner
Acs Omega, 7, 2022
|
|
8Y7L
 
 | |
5EA0
 
 | | Structure of the antibody 7968 with human complement factor H-derived peptide | | Descriptor: | Complement factor H-related protein 2, Heavy chain of antibody 7968 Fab fragment, Light chain of antibody 7968 Fab fragment | | Authors: | Bushey, R.T, Moody, M.A, Nicely, N.I, Alam, S.M, Haynes, B.F, Winkler, M.T, Gottlin, E.B, Campa, M.J, Liao, H.-X, Patz Jr, E.F. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Therapeutic Antibody for Cancer, Derived from Single Human B Cells.
Cell Rep, 15, 2016
|
|
3F0Y
 
 | | Crystal structure of the human Adenovirus type 14 fiber knob | | Descriptor: | Fiber protein, GLYCEROL, IMIDAZOLE | | Authors: | Persson, B.D, Reiter, D.M, Arnberg, N, Stehle, T. | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An arginine switch in the species B adenovirus knob determines high-affinity engagement of cellular receptor CD46
J.Virol., 83, 2009
|
|
7U0D
 
 | |
