6BSE
 
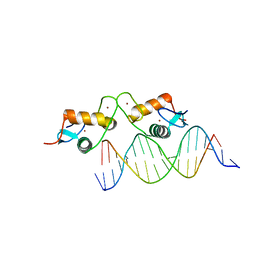 | | Glucocorticoid receptor bound to high cooperativity monomer sequence | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*CP*GP*TP*GP*TP*AP*CP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*AP*AP*AP*AP*AP*GP*TP*AP*CP*AP*CP*GP*TP*GP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | General and sequence-specific roles for DNA in glucocorticoid receptor DNA-binding stoichiometry
To Be Published
|
|
6BSQ
 
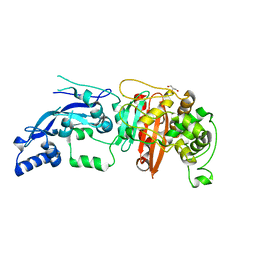 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | Descriptor: | CHLORIDE ION, GLYCEROL, PBP4 protein | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6C9U
 
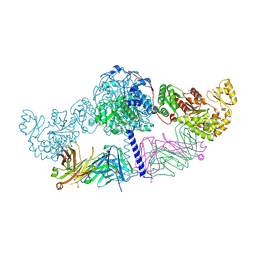 | | Crystal structure of [KS3][AT3] didomain from module 3 of 6-deoxyerthronolide B synthase in complex with antibody fragment (Fab) | | Descriptor: | 6-deoxyerythronolide-B synthase EryA2, modules 3 and 4, Heavy chain of Fab 1B2, ... | | Authors: | Deis, L.N, Li, X, Mathews, I.I, Khosla, C. | | Deposit date: | 2018-01-28 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Function Analysis of the Extended Conformation of a Polyketide Synthase Module.
J. Am. Chem. Soc., 140, 2018
|
|
6CAA
 
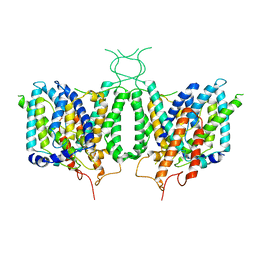 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
6CAV
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia in complex with GMPPNP, Magnesium, and Dibekacin | | Descriptor: | Bifunctional AAC/APH, CHLORIDE ION, Dibekacin, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2018-02-01 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasticity of Aminoglycoside Binding to Antibiotic Kinase APH(2′′)-Ia.
Antimicrob. Agents Chemother., 62, 2018
|
|
6BT1
 
 | | Structure of the human Nocturnin catalytic domain | | Descriptor: | MAGNESIUM ION, Nocturnin | | Authors: | Abshire, E.T, Chasseur, J, Del Rizzo, P, Trievel, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The structure of human Nocturnin reveals a conserved ribonuclease domain that represses target transcript translation and abundance in cells.
Nucleic Acids Res., 46, 2018
|
|
6BTA
 
 | | CypA Mutant - S99T C115S | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Fraser, J.S, Kenner, L.R, Liu, L. | | Deposit date: | 2017-12-06 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescue of conformational dynamics in enzyme catalysis by directed evolution.
Nat Commun, 9, 2018
|
|
6BTM
 
 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | Authors: | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | Deposit date: | 2017-12-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
6CB8
 
 | |
6BUQ
 
 | |
1Y52
 
 | | structure of insect cell (Baculovirus) expressed AVR4 (C122S)-biotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin-related protein 4/5, BIOTIN | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6BWK
 
 | |
6BR9
 
 | | Structure of A6 reveals a novel lipid transporter | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Protein A6 homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, J, Peng, S, Pathak, P. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BRG
 
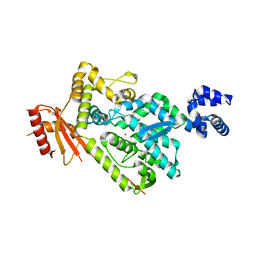 | | The SAM domain of mouse SAMHD1 is critical for its activation and regulation | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Buzovetsky, O, Tang, C, Knecht, K.M, Antonucci, J.M, Wu, L, Ji, X, Xiong, Y. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The SAM domain of mouse SAMHD1 is critical for its activation and regulation.
Nat Commun, 9, 2018
|
|
6BWP
 
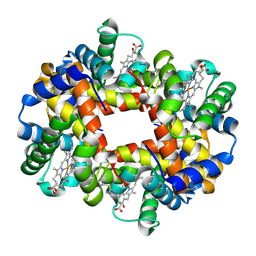 | | Crystal structure of Deoxy Hemoglobin in complex with beta Cys93 modifying agent, TD3 | | Descriptor: | 1H-1,2,3-triazole-5-thiol, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Musayev, F.N, Safo, R.M, Safo, M.K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Triazole Disulfide Compound Increases the Affinity of Hemoglobin for Oxygen and Reduces the Sickling of Human Sickle Cells.
Mol. Pharm., 15, 2018
|
|
1KME
 
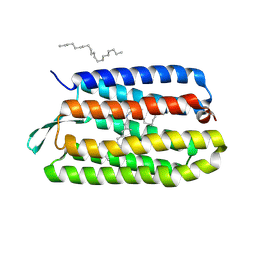 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN CRYSTALLIZED FROM BICELLES | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, Bacteriorhodopsin, RETINAL, ... | | Authors: | Faham, S, Bowie, J.U. | | Deposit date: | 2001-12-14 | | Release date: | 2002-02-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure.
J.Mol.Biol., 316, 2002
|
|
6BT3
 
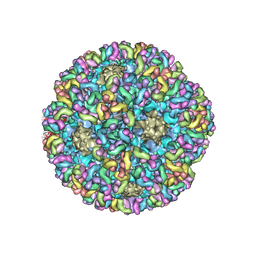 | | High-Resolution Structure Analysis of Antibody V5 Conformational Epitope on Human Papillomavirus 16 | | Descriptor: | Major capsid protein L1, V5 Fab Heavy-chain, V5 Fab Light-chain | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Ashley, R.E, Makhov, A.M, Conway, J.F, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2017-12-05 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16.
Viruses, 9, 2017
|
|
6BUZ
 
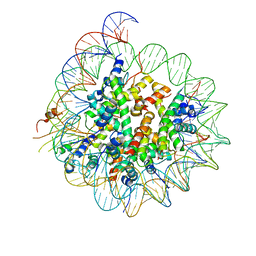 | | Cryo-EM structure of CENP-A nucleosome in complex with kinetochore protein CENP-N | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Chittori, S, Hong, J, Kelly, A.E, Bai, Y, Subramaniam, S. | | Deposit date: | 2017-12-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N.
Science, 359, 2018
|
|
5HWU
 
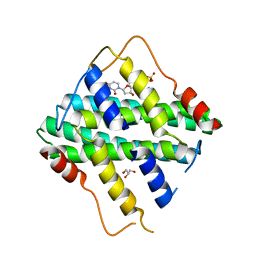 | | Crystal Structure of DR2231_E46A mutant in complex with dUMPNPP and Manganese | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DR2231, GLYCEROL, ... | | Authors: | Mota, C.S, Goncalves, A.M.D, de Sanctis, D. | | Deposit date: | 2016-01-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deinococcus radiodurans DR2231 is a two-metal-ion mechanism hydrolase with exclusive activity on dUTP.
FEBS J., 283, 2016
|
|
6BVD
 
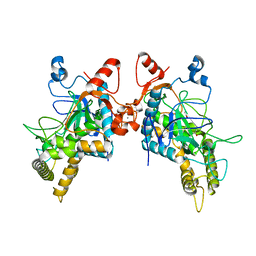 | | Structure of Botulinum Neurotoxin Serotype HA Light Chain | | Descriptor: | ACETATE ION, CALCIUM ION, Light Chain, ... | | Authors: | Jin, R, Lam, K. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical characterization of the protease domain of the mosaic botulinum neurotoxin type HA.
Pathog Dis, 76, 2018
|
|
6BW2
 
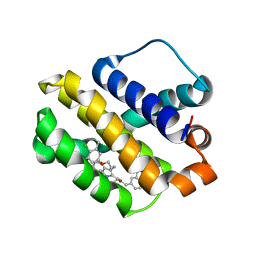 | | Mcl-1 complexed with small molecules | | Descriptor: | 3-({11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}methyl)benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of Potent and Selective Tricyclic Indole Diazepinone Myeloid Cell Leukemia-1 Inhibitors Using Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6BWG
 
 | |
1YAH
 
 | | Crystal Structure of Human Liver Carboxylesterase complexed to Etyl Acetate; A Fatty Acid Ethyl Ester Analogue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CES1 protein, ETHYL ACETATE, ... | | Authors: | Fleming, C.D, Bencahrit, S, Edwards, C.C, Hyatt, J.L, Morton, C.L, Howard-Williams, E.L, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2004-12-17 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into drug processing by human carboxylesterase 1: tamoxifen, mevastatin, and inhibition by benzil.
J.Mol.Biol., 352, 2005
|
|
6BWV
 
 | | Crystal Structure of the 4-1BB/4-1BBL Complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Oganesyan, V, Gilbreth, R.N, Baca, M. | | Deposit date: | 2017-12-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human 4-1BB/4-1BBL complex.
J. Biol. Chem., 293, 2018
|
|
6U7N
 
 | | Crystal structure of neurotrimin (NTM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Neurotrimin, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G, Rush, S. | | Deposit date: | 2019-09-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
