5P3K
 
 | |
5P3W
 
 | |
5P4C
 
 | |
5P4R
 
 | |
5P55
 
 | |
5P5K
 
 | |
5P60
 
 | |
7RB7
 
 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA and MMB4 oxime | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
7RB5
 
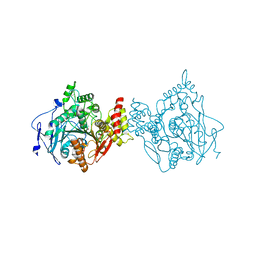 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA | | Descriptor: | 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
7RB6
 
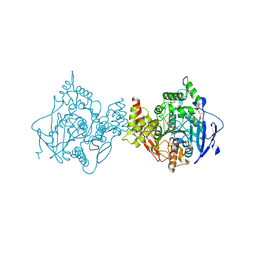 | | Low temperature structure of hAChE in complex with substrate analog 4K-TMA | | Descriptor: | 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
3ZU7
 
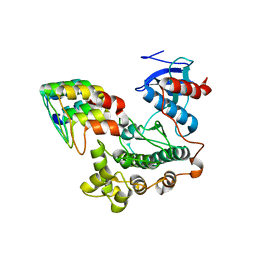 | |
3ZUV
 
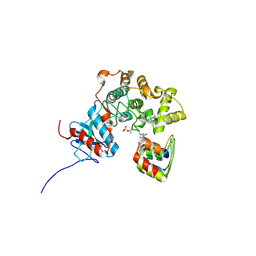 | |
5P0J
 
 | |
5P0X
 
 | |
5P1B
 
 | |
5P1M
 
 | |
5P1W
 
 | |
5P2A
 
 | |
5OT3
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp191 and RKp117 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, [2-[(4-isoquinolin-5-ylsulfonyl-1,4-diazepan-1-yl)methyl]phenyl]boronic acid, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-08-19 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
5P2O
 
 | |
5P31
 
 | |
5P3H
 
 | |
5P3Y
 
 | |
5P4D
 
 | |
5P4S
 
 | |
