5DTB
 
 | |
5E9R
 
 | |
2BQJ
 
 | |
2BQA
 
 | |
2BQC
 
 | |
2BQB
 
 | |
4UD9
 
 | | Thrombin in complex with 5-chlorothiophene-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-CHLORO-2-THIOPHENECARBOXAMIDE, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2014-12-09 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
2BQE
 
 | |
2BQO
 
 | |
2BQF
 
 | |
2BQN
 
 | |
5EBA
 
 | |
6S09
 
 | | C-terminally extended and N-terminally truncated variant of FimA E. coli at 1.5 Angstrom resolution | | Descriptor: | ACETIC ACID, FimA, SODIUM ION, ... | | Authors: | Zyla, D, Echeverria, B, Glockshuber, R. | | Deposit date: | 2019-06-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Donor strand sequence, rather than donor strand orientation, determines the stability and non-equilibrium folding of the type 1 pilus subunit FimA.
J.Biol.Chem., 295, 2020
|
|
6A38
 
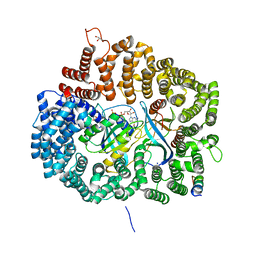 | | MVM NS2 NES in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, Exportin-1, GLYCEROL, ... | | Authors: | Sun, Q, Li, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Cancer Therapy with Nanoparticle-Medicated Intracellular Expression of Peptide CRM1-Inhibitor.
Int J Nanomedicine, 16, 2021
|
|
6A47
 
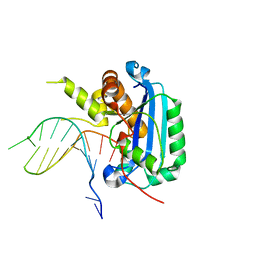 | | Structure of TREX2 in complex with a Y structured dsDNA | | Descriptor: | DNA (5'-D(P*CP*CP*AP*GP*GP*CP*CP*CP*TP*CP*TP*AP*GP*GP*GP*CP*CP*TP*T)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Hsiao, Y.Y. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the duplex DNA processing of TREX2
Nucleic Acids Res., 46, 2018
|
|
7Z50
 
 | |
6A4N
 
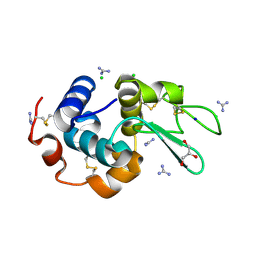 | | HEWL crystals soaked in 2.5M GuHCl for 8 minutes | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANIDINE, ... | | Authors: | Tushar, R, Kini, R.M, Koh, C.Y, Hosur, M.V. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystallographic analysis of time-dependent binding of guanidine hydrochloride to HEWL: First steps during protein unfolding.
Int. J. Biol. Macromol., 122, 2019
|
|
6RNL
 
 | | L-[Ru(TAP)2(dppz)]2+ bound to the G-quadruplex forming sequence d(TAGGGTT) | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*TP*T)-3'), POTASSIUM ION, Ru(tap)2(dppz) complex, ... | | Authors: | McQuaid, K.T, Hall, J.P, Cardin, C.J. | | Deposit date: | 2019-05-09 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Three thymine/adenine binding modes of the ruthenium complex Lambda-[Ru(TAP)2(dppz)]2+to the G-quadruplex forming sequence d(TAGGGTT) shown by X-ray crystallography.
Chem.Commun.(Camb.), 55, 2019
|
|
6A83
 
 | |
4UEH
 
 | | Thrombin in complex with benzamidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2014-12-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
4UE7
 
 | | Thrombin in complex with 1-amidinopiperidine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2014-12-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.129 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
4UFF
 
 | | Thrombin in complex with (2R)-2-(benzylsulfonylamino)-N-(2-((4- carbamimidoylphenyl)methylamino)-2-oxo-ethyl)-N-methyl-3-phenyl- propanamide | | Descriptor: | (2R)-2-(benzylsulfonylamino)-N-(2-((4-carbamimidoylphenyl)methylamino)-2-oxo-ethyl)-N-methyl-3-phenyl-propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-2, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Boosting Affinity by Correct Ligand Preorganization for the S2 Pocket of Thrombin: A Study by Isothermal Titration Calorimetry, Molecular Dynamics, and High-Resolution Crystal Structures.
Chemmedchem, 11, 2016
|
|
6RU6
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with monophosphorylated p63 PAD1P peptide | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6S16
 
 | | T. thermophilus RuvC in complex with Holliday junction substrate | | Descriptor: | CHLORIDE ION, Crossover junction endodeoxyribonuclease RuvC, DNA (33-MER), ... | | Authors: | Gorecka, K.M, Krepl, M, Szlachcic, A, Poznanski, J, Sponer, J, Nowotny, M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | RuvC uses dynamic probing of the Holliday junction to achieve sequence specificity and efficient resolution.
Nat Commun, 10, 2019
|
|
4UFD
 
 | | Thrombin in complex with 4-(((1-((2S)-1-((2R)-2-(benzylsulfonylamino)- 3-phenyl-propanoyl)pyrrolidin-2-yl)-1-oxo-ethyl)amino)methyl) benzamidine | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-1-[(2R)-3-phenyl-2-[(phenylmethyl)sulfonylamino]propanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Boosting Affinity by Correct Ligand Preorganization for the S2 Pocket of Thrombin: A Study by Isothermal Titration Calorimetry, Molecular Dynamics, and High-Resolution Crystal Structures.
Chemmedchem, 11, 2016
|
|
