6MVJ
 
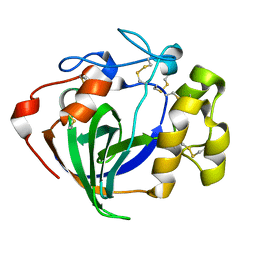 | | Cellobiose complex Cel45A from Neurospora crassa OR74A | | Descriptor: | Endoglucanase V, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | Structural insights into the hydrolysis pattern and molecular dynamics simulations of GH45 subfamily a endoglucanase from Neurospora crassa OR74A.
Biochimie, 165, 2019
|
|
8E9I
 
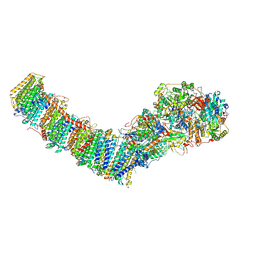 | | Mycobacterial respiratory complex I, semi-inserted quinone | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E26
 
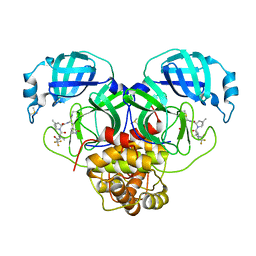 | | Crystal Structure of SARS-CoV-2 Main Protease N142S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-08-14 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
1XEC
 
 | | Dimeric bovine tissue-extracted decorin, crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Decorin | | Authors: | Scott, P.G, McEwan, P.A, Dodd, C.M, Bergmann, E.M, Bishop, P.N, Bella, J. | | Deposit date: | 2004-09-09 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2J15
 
 | | Cyclic MrIA: An exceptionally stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter. | | Descriptor: | MAI126P | | Authors: | Lovelace, E.S, Armishaw, C.J, Colgrave, M.L, Walstrom, M.E, Alewood, P.F, Daly, N.L, Craik, D.J. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-01 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Cyclic MrIA: a stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter.
J. Med. Chem., 49, 2006
|
|
2YLP
 
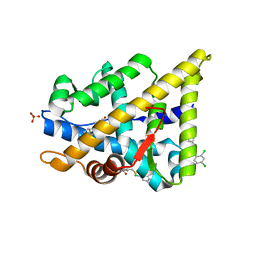 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 3-[(2,4-DICHLOROPHENYL)METHYLSULFANYLMETHYL]BENZOIC ACID, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
8E9H
 
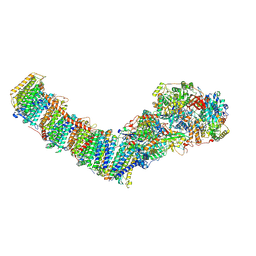 | | Mycobacterial respiratory complex I, fully-inserted quinone | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2J75
 
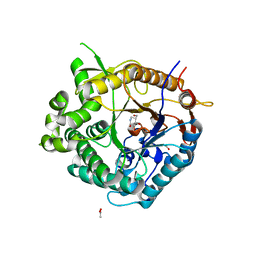 | | Beta-glucosidase from Thermotoga maritima in complex with noeuromycin | | Descriptor: | (2R,3S,4R,5R)-5-(HYDROXYMETHYL)PIPERIDINE-2,3,4-TRIOL, ACETATE ION, BETA-GLUCOSIDASE A, ... | | Authors: | Gloster, T.M, Meloncelli, P, Stick, R.V, Zechel, D, Davies, G.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
1XM4
 
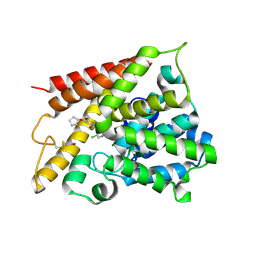 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Piclamilast | | Descriptor: | 3-(CYCLOPENTYLOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-METHOXYBENZAMIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-01 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
2YE9
 
 | |
7F7E
 
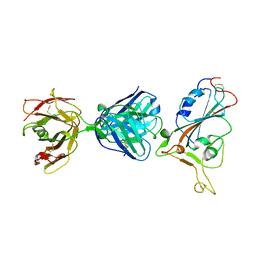 | | SARS-CoV-2 S protein RBD in complex with A5-10 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A5-10 Fab, Light chain of A5-10 Fab, ... | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Etesevimab in combination with JS026 neutralizing SARS-CoV-2 and its variants.
Emerg Microbes Infect, 11, 2022
|
|
8E9G
 
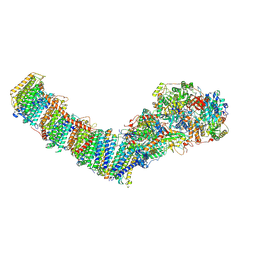 | | Mycobacterial respiratory complex I with both quinone positions modelled | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6MX6
 
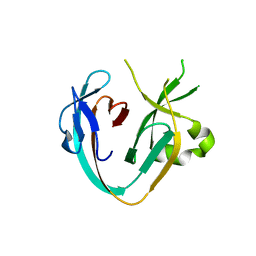 | | The Prp8 intein of Cryptococcus neoformans | | Descriptor: | Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Li, H. | | Deposit date: | 2018-10-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Spliceosomal Prp8 intein at the crossroads of protein and RNA splicing.
Plos Biol., 17, 2019
|
|
6MXG
 
 | |
6MXT
 
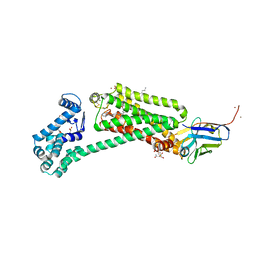 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
3L54
 
 | | Structure of Pi3K gamma with inhibitor | | Descriptor: | 6-(1H-pyrazolo[3,4-b]pyridin-5-yl)-4-pyridin-4-ylquinoline, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Elkins, P.A, Smallwood, A.M. | | Deposit date: | 2009-12-21 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of GSK2126458, a Highly Potent Inhibitor of PI3K and the Mammalian Target of Rapamycin.
ACS Med Chem Lett, 1, 2010
|
|
2YI0
 
 | | Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. | | Descriptor: | 4-CHLORO-6-[5-(4-METHOXYPHENYL)-1,2,3-THIADIAZOL-4-YL]BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-05-10 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-Crystalization and in Vitro Biological Characterization of 5-Aryl-4-(5-Substituted-2-4-Dihydroxyphenyl)-1,2,3-Thiadiazole Hsp90 Inhibitors.
Plos One, 7, 2012
|
|
8E25
 
 | | Crystal Structure of SARS-CoV-2 Main Protease M49I mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-08-14 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
2YHV
 
 | |
3KDU
 
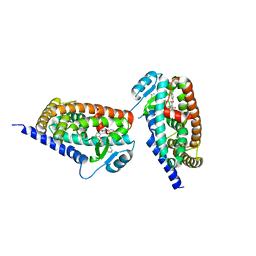 | |
2YEF
 
 | |
2YEB
 
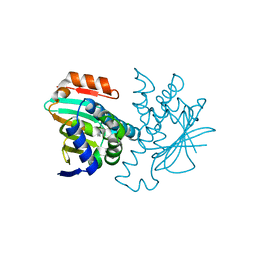 | |
4R2U
 
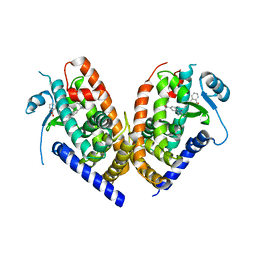 | | Crystal Structure of PPARgamma in complex with SR1664 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1S)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Marciano, D.P, Kamenecka, T, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-08-13 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological repression of PPAR gamma promotes osteogenesis.
Nat Commun, 6, 2015
|
|
2JLE
 
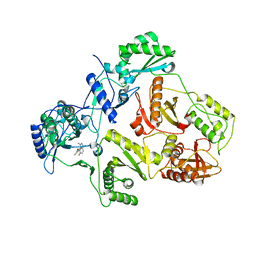 | | Novel indazole nnrtis created using molecular template hybridization based on crystallographic overlays | | Descriptor: | 5-[(5-fluoro-3-methyl-1H-indazol-4-yl)oxy]benzene-1,3-dicarbonitrile, REVERSE TRANSCRIPTASE/RNASEH | | Authors: | Jones, L.H, Allan, G, Barba, O, Burt, C, Corbau, R, Dupont, T, Irving, S, Mowbray, C.E, Phillips, C, Swain, N.A, Webster, R, Westby, M. | | Deposit date: | 2008-09-08 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Indazole Non-Nucleoside Reverse Transcriptase Inhibitors Using Molecular Hybridization Based on Crystallographic Overlays.
J.Med.Chem., 52, 2009
|
|
4R17
 
 | | Ligand-induced aziridine-formation at subunit beta5 of the yeast 20S proteasome | | Descriptor: | (2S,3S)-3-methylaziridine-2-carboxylic acid, MAGNESIUM ION, Proteasome subunit alpha type-1, ... | | Authors: | Dubiella, C, Cui, H, Gersch, M, Brouwer, A.J, Sieber, S.A, Krueger, A, Liskamp, R, Groll, M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective inhibition of the immunoproteasome by ligand-induced crosslinking of the active site.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
