3F4E
 
 | |
3F4F
 
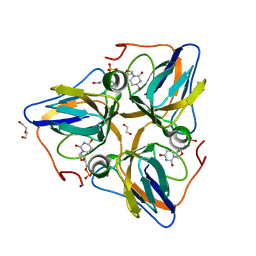 | | Crystal structure of dUT1p, a dUTPase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3F4G
 
 | |
3F4H
 
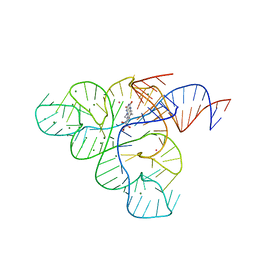
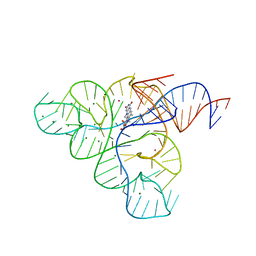 | | Crystal structure of the FMN riboswitch bound to roseoflavin | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, FMN riboswitch, MAGNESIUM ION, ... | | Authors: | Serganov, A.A, Huang, L. | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch.
Nature, 458, 2009
|
|
3F4I
 
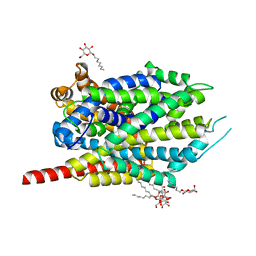 | | Crystal Structure of LeuT bound to L-selenomethionine and sodium | | Descriptor: | SELENOMETHIONINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-31 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3F4J
 
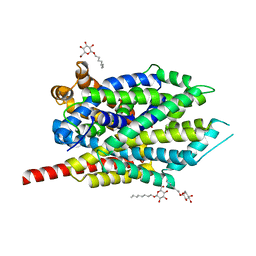 | | Crystal structure of LeuT bound to glycine and sodium | | Descriptor: | GLYCINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-31 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3F4K
 
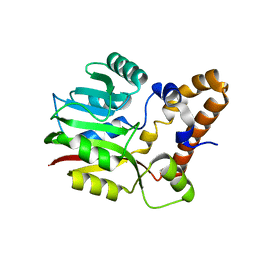 | | Crystal structure of a probable methyltransferase from Bacteroides thetaiotaomicron. Northeast Structural Genomics target BtR309. | | Descriptor: | Putative methyltransferase | | Authors: | Seetharaman, J, Lew, S, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a probable methyltransferase from Bacteroides thetaiotaomicron. Northeast Structural Genomics target BtR309.
To be Published
|
|
3F4L
 
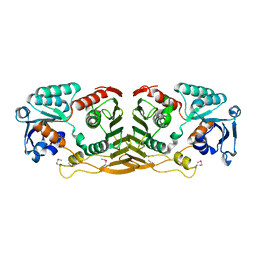 | | Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 | | Descriptor: | Putative oxidoreductase yhhX | | Authors: | Seetharaman, J, Abashidze, M, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: |
|
|
3F4M
 
 | | Crystal structure of TIPE2 | | Descriptor: | CHLORIDE ION, Tumor necrosis factor, alpha-induced protein 8-like protein 2 | | Authors: | Zhang, X, Wang, J, Shi, Y. | | Deposit date: | 2008-11-01 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Crystal structure of TIPE2 provides insights into immune homeostasis
Nat.Struct.Mol.Biol., 16, 2009
|
|
3F4N
 
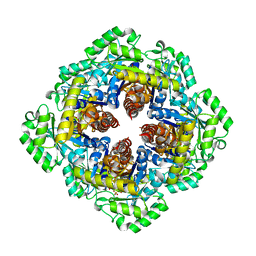 | | Crystal Structure of Pyridoxal Phosphate Biosynthetic Protein PdxJ from Yersinia pestis | | Descriptor: | PYRIDOXINE-5'-PHOSPHATE, Pyridoxine 5'-phosphate synthase, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-11-01 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal Structure of Pyridoxal Phosphate Biosynthetic Protein PdxJ from Yersinia pestis
To be Published, 2008
|
|
3F4R
 
 | | Crystal structure of Wolbachia pipientis alpha-DsbA1 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative uncharacterized protein, TRIETHYLENE GLYCOL | | Authors: | Kurz, M, Heras, B, Martin, J.L. | | Deposit date: | 2008-11-02 | | Release date: | 2009-03-24 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the Oxidoreductase alpha-DsbA1 from Wolbachia pipientis.
ANTIOXID.REDOX SIGNAL., 11, 2009
|
|
3F4S
 
 | |
3F4T
 
 | |
3F4V
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F4W
 
 | |
3F4X
 
 | | Carbonic anhydrase inhibitors. Comparison of chlorthalidone and indapamide X-ray crystal structures in adducts with isozyme II: when three water molecules make the difference | | Descriptor: | 2-chloro-5-[(1S)-1-hydroxy-3-oxo-2H-isoindol-1-yl]benzenesulfonamide, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Temperini, C, Cecchi, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2008-11-03 | | Release date: | 2009-03-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase inhibitors. Comparison of chlorthalidone and indapamide X-ray crystal structures in adducts with isozyme II: when three water molecules and the keto-enol tautomerism make the difference.
J.Med.Chem., 52, 2009
|
|
3F4Y
 
 | |
3F4Z
 
 | |
3F50
 
 | |
3F51
 
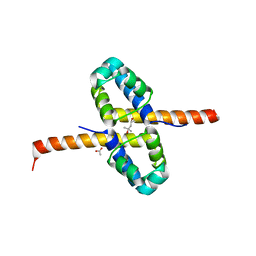 | | Crystal Structure of the clp gene regulator ClgR from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
3F52
 
 | | Crystal structure of the clp gene regulator ClgR from C. glutamicum | | Descriptor: | GLYCEROL, clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
3F53
 
 | |
3F56
 
 | |
3F57
 
 | |
3F58
 
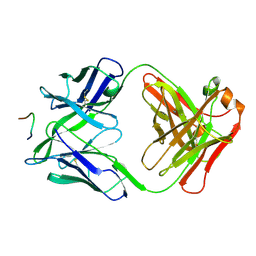 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 12-RESIDUE CYCLIC PEPTIDE (INCLUDING RESIDUES 315-324 OF HIV-1 GP120 (MN ISOLATE); H315S MUTATION | | Descriptor: | PROTEIN (CYCLIC PEPTIDE (GP120)), PROTEIN (IMMUNOGLOBULIN GAMMA I (58.2)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-23 | | Release date: | 1999-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
