1T0G
 
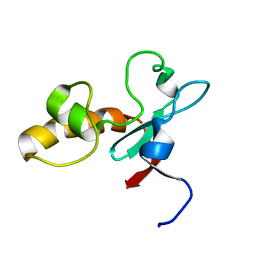 | | Hypothetical protein At2g24940.1 from Arabidopsis thaliana has a cytochrome b5 like fold | | Descriptor: | cytochrome b5 domain-containing protein | | Authors: | Song, J, Vinarov, D.A, Tyler, E.M, Shahan, M.N, Tyler, R.C, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-08 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hypothetical protein At2g24940.1 from Arabidopsis thaliana has a cytochrome b5 like fold
J.Biomol.NMR, 30, 2004
|
|
4GG1
 
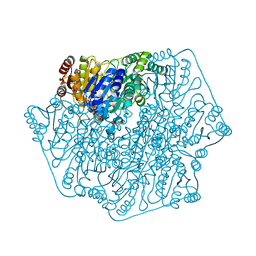 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403T | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-04 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
4OWB
 
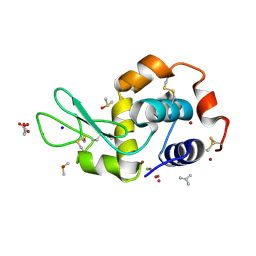 | |
1T4R
 
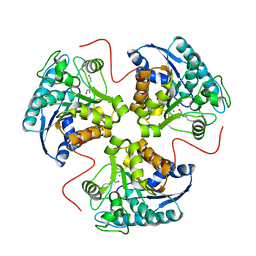 | | arginase-descarboxy-nor-NOHA complex | | Descriptor: | 3-{[(E)-AMINO(HYDROXYIMINO)METHYL]AMINO}PROPAN-1-AMINIUM, Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
7GAW
 
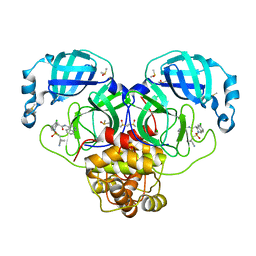 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-e194df51-1 (SARS2_MproA-x0862) | | Descriptor: | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4FUY
 
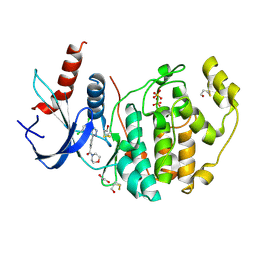 | |
5RHP
 
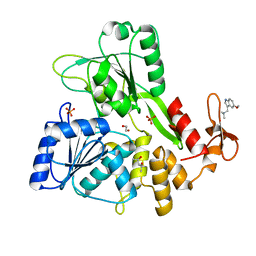 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z2856434938 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3RJT
 
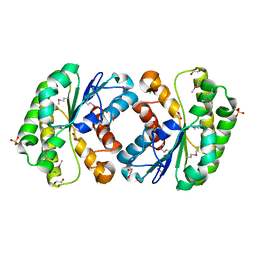 | | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Chang, C, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-15 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
7GBB
 
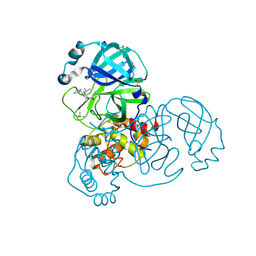 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAN-LON-a5fc619e-3 (Mpro-x10306) | | Descriptor: | 1-[(3S)-4-[(3-chlorophenyl)methyl]-3-(2-methylpropyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2H21
 
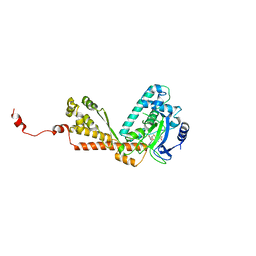 | | Structure of Rubisco LSMT bound to AdoMet | | Descriptor: | Ribulose-1,5 bisphosphate carboxylase/oxygenase, S-ADENOSYLMETHIONINE | | Authors: | Couture, J.F, Hauk, G, Trievel, R.C. | | Deposit date: | 2006-05-17 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Catalytic Roles for Carbon-Oxygen Hydrogen Bonding in SET Domain Lysine Methyltransferases.
J.Biol.Chem., 281, 2006
|
|
4J8Q
 
 | |
3RNJ
 
 | | Crystal structure of the SH3 domain from IRSp53 (BAIAP2) | | Descriptor: | 1,2-ETHANEDIOL, Brain-specific angiogenesis inhibitor 1-associated protein 2, ISOPROPYL ALCOHOL, ... | | Authors: | Simister, P.C, Barilari, M, Muniz, J.R.C, Dente, L, Knapp, S, von Delft, F, Filippakopoulos, P, Vollmar, M, Chaikuad, A, Raynor, J, Tregubova, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the SH3 domain from IRSp53 (BAIAP2)
To be Published
|
|
3R1X
 
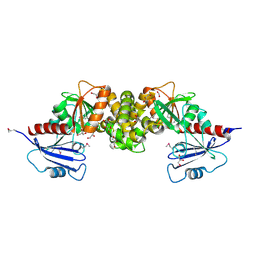 | | Crystal structure of 2-oxo-3-deoxygalactonate kinase from Klebsiella pneumoniae | | Descriptor: | 2-oxo-3-deoxygalactonate kinase, FORMIC ACID, GLYCEROL | | Authors: | Michalska, K, Cuff, M.E, Tesar, C, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-11 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structure of 2-oxo-3-deoxygalactonate kinase from Klebsiella pneumoniae.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1DK6
 
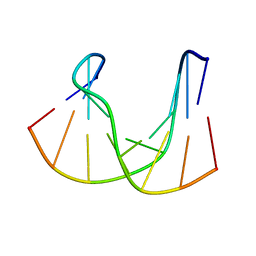 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 2000-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
3O7S
 
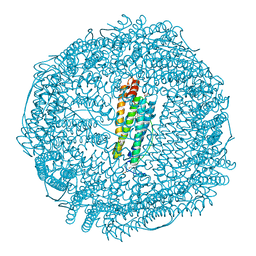 | | Crystal structure of Ru(p-cymene)/apo-Fr | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4-(1-methylethyl)benzene, CADMIUM ION, ... | | Authors: | Takezawa, Y, Bockmann, P, Sugi, N, Wang, Z, Abe, S, Murakami, T, Hikage, T, Erker, G, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-07-31 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Incorporation of organometallic Ru complexes into apo-ferritin cage.
J.Chem.Soc.,Dalton Trans., 40, 2011
|
|
3RBM
 
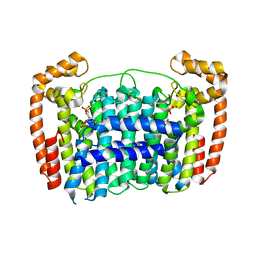 | | Crystal structure of Plasmodium vivax geranylgeranylpyrophosphate synthase complexed with BPH -703 | | Descriptor: | 3-(2,2-diphosphonoethyl)-1-dodecyl-1H-imidazol-3-ium, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Liu, Y.L, No, J.H, Oldfield, E. | | Deposit date: | 2011-03-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Lipophilic analogs of zoledronate and risedronate inhibit Plasmodium geranylgeranyl diphosphate synthase (GGPPS) and exhibit potent antimalarial activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7FTM
 
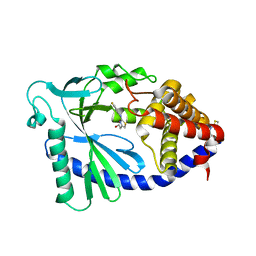 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[2-(4-fluoroanilino)-1,3-thiazol-4-yl]acetic acid | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, [2-(4-fluoroanilino)-1,3-thiazol-4-yl]acetic acid | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
5RWG
 
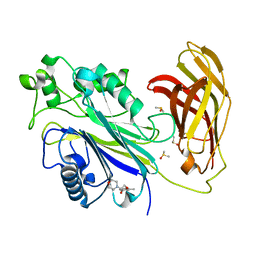 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z17497990 | | Descriptor: | 2-(2,4-dimethylphenoxy)-1-morpholin-4-yl-ethanone, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
3RDA
 
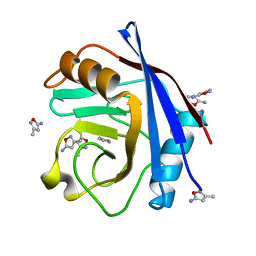 | | Human Cyclophilin D Complexed with a Fragment | | Descriptor: | 3-methyl-1,2-oxazol-5-amine, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-04-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
5RXJ
 
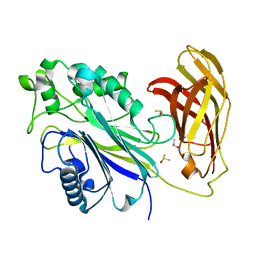 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1722836661 | | Descriptor: | 2-(1H-imidazol-1-yl)ethyl dimethylcarbamate, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
1XJO
 
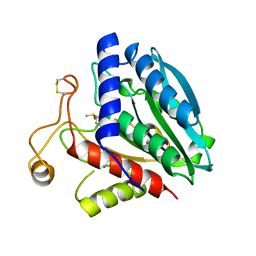 | | STRUCTURE OF AMINOPEPTIDASE | | Descriptor: | AMINOPEPTIDASE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Greenblatt, H.M, Barra, D, Blumberg, S, Shoham, G. | | Deposit date: | 1996-10-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Streptomyces griseus aminopeptidase: X-ray crystallographic structure at 1.75 A resolution.
J.Mol.Biol., 265, 1997
|
|
7FTR
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with (Z)-N-(4-acetylphenyl)-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide | | Descriptor: | (2Z)-N-(4-acetylphenyl)-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide, ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FTS
 
 | |
5RZ2
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z68277692 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
7GAL
 
 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z291279160 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(trifluoromethyloxy)phenyl]thiourea, CHLORIDE ION, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
