2R6S
 
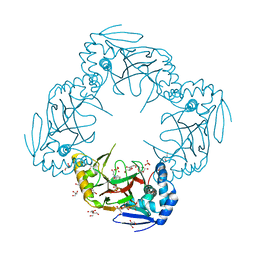 | | Crystal structure of Gab protein | | Descriptor: | BICINE, FE (II) ION, GLYCEROL, ... | | Authors: | Lohkamp, B, Dobritzsch, D. | | Deposit date: | 2007-09-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mixture of fortunes: the curious determination of the structure of Escherichia coli BL21 Gab protein.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2RIS
 
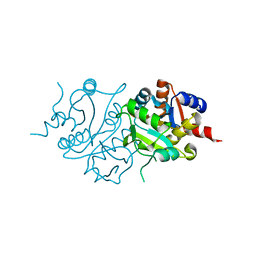 | |
6UX5
 
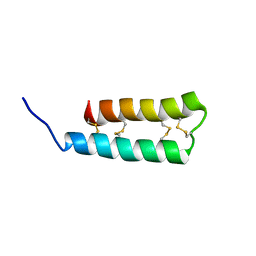 | |
2W3B
 
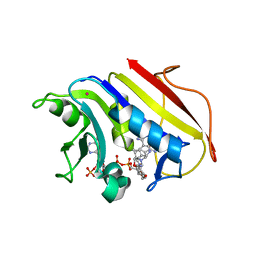 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND A LIPOPHILIC ANTIFOLATE SELECTIVE FOR MYCOBACTERIUM AVIUM DHFR, 6-((2,5- DIETHOXYPHENYL)AMINOMETHYL)-2,4-DIAMINO-5-METHYLPYRIDO(2,3-D) PYRIMIDINE (SRI-8686) | | Descriptor: | 6-{[(2,5-DIETHOXYPHENYL)AMINO]METHYL}-5-METHYLPYRIDO[2,3-D]PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE, FOLIC ACID, ... | | Authors: | Leung, A.K.W, Reynolds, R.C, Borhani, D.W. | | Deposit date: | 2008-11-11 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural Basis for Selective Inhibition of Mycobacterium Avium Dihydrofolate Reductase by a Lipophilic Antifolate
To be Published
|
|
8C3T
 
 | |
6VPB
 
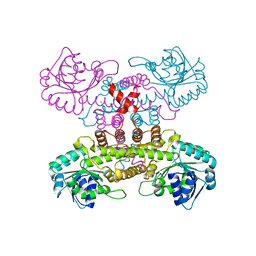 | |
6KTQ
 
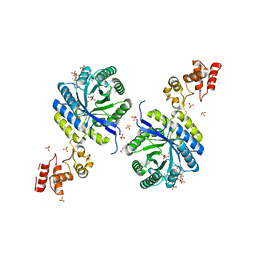 | | Crystal structure of catalytic domain of homocitrate synthase from Sulfolobus acidocaldarius (SaHCS(dRAM)) in complex with alpha-ketoglutarate/Zn2+/CoA | | Descriptor: | 2-OXOGLUTARIC ACID, COENZYME A, Homocitrate synthase, ... | | Authors: | Suzuki, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Involvement of subdomain II in the recognition of acetyl-CoA revealed by the crystal structure of homocitrate synthase from Sulfolobus acidocaldarius.
Febs J., 288, 2021
|
|
8R0I
 
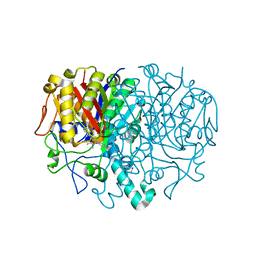 | | Pseudomonas aeruginosa FabF C164A in complex with 3-amino-N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)benzamide | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-N-(1,5-dimethyl-3-oxidanylidene-2-phenyl-pyrazol-4-yl)benzamide, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Yadrykhinsky, V, Brenk, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
4PCA
 
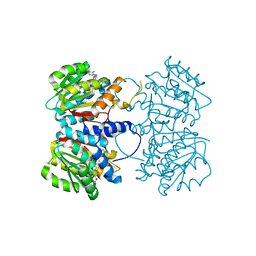 | | X-ray crystal structure of an O-methyltransferase from Anaplasma phagocytophilum bound to SAH and Manganese | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, O-methyltransferase family protein, ... | | Authors: | Fairman, J.W, Abendroth, J, Lorimer, D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An O-Methyltransferase Is Required for Infection of Tick Cells by Anaplasma phagocytophilum.
Plos Pathog., 11, 2015
|
|
8R1V
 
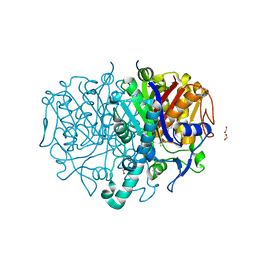 | | Pseudomonas aeruginosa FabF C164A in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-2-(4-methoxyphenoxy)acetamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Yadrykhinsky, V, Brenk, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
5MJG
 
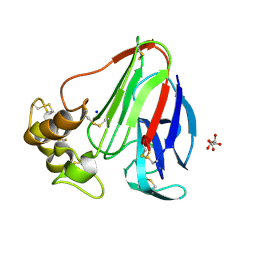 | | Single-shot pink beam serial crystallography: Thaumatin | | Descriptor: | S,R MESO-TARTARIC ACID, SODIUM ION, Thaumatin-1 | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-shot pink beam serial crystallography: Thaumatin
To Be Published
|
|
2W3A
 
 | |
5DOW
 
 | |
4PCL
 
 | | X-ray crystal structure of an O-methyltransferase from Anaplasma phagocytophilum bound to SAM and a Manganese ion. | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, O-methyltransferase family protein, ... | | Authors: | Fairman, J.W, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-04-15 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An O-Methyltransferase Is Required for Infection of Tick Cells by Anaplasma phagocytophilum.
Plos Pathog., 11, 2015
|
|
8DTA
 
 | |
6MBE
 
 | | Human Mcl-1 in complex with the designed peptide dM7 | | Descriptor: | CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, dM7 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
9PF2
 
 | | NSF, substrate free, hydrolyzing, class 25 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | White, K.I, Brunger, A.T. | | Deposit date: | 2025-07-03 | | Release date: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural remodeling of target-SNARE protein complexes by NSF enables synaptic transmission.
Nat Commun, 16, 2025
|
|
9PFF
 
 | | Min22bin20S complex (NSF-alphaSNAP-2:2 syntaxin-1a H3:SNAP-25 SN1), non-hydrolyzing, class 27 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Brunger, A.T. | | Deposit date: | 2025-07-04 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural remodeling of target-SNARE protein complexes by NSF enables synaptic transmission.
Nat Commun, 16, 2025
|
|
9PFG
 
 | | Min22bin20S complex (NSF-alphaSNAP-2:2 syntaxin-1a H3:SNAP-25 SN1), 4:2:2 alphaSNAP-syntaxin-1a H3-SNAP-25 SN1 subcomplex local refinement, non-hydrolyzing, class 28 | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | White, K.I, Brunger, A.T. | | Deposit date: | 2025-07-04 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural remodeling of target-SNARE protein complexes by NSF enables synaptic transmission.
Nat Commun, 16, 2025
|
|
6Q2F
 
 | | Structure of Rhamnosidase from Novosphingobium sp. PP1Y | | Descriptor: | Glycoside hydrolase family protein, SODIUM ION | | Authors: | Terry, B, Ha, J, Izzo, V, Sazinsky, M.H. | | Deposit date: | 2019-08-07 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.20000076 Å) | | Cite: | The crystal structure and insight into the substrate specificity of the alpha-L rhamnosidase RHA-P from Novosphingobium sp. PP1Y.
Arch.Biochem.Biophys., 679, 2019
|
|
6GIM
 
 | | Structure of the DNA duplex d(AAATTT)2 with [N-(3-chloro-4-((4,5-dihydro-1H-imidazol-2-yl)amino)phenyl)-4-((4,5-dihydro-1H-imidazol-2- yl)amino)benzamide] - (drug JNI18) | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*T)-3'), MAGNESIUM ION, [4-[(3-chloranyl-4-imidazolidin-2-ylideneazaniumyl-phenyl)carbamoyl]phenyl]-imidazolidin-2-ylidene-azanium | | Authors: | Millan, C.R, Dardonvile, C, de Koning, H.P, Saperas, N, Campos, J.L. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Functional and structural analysis of AT-specific minor groove binders that disrupt DNA-protein interactions and cause disintegration of the Trypanosoma brucei kinetoplast.
Nucleic Acids Res., 45, 2017
|
|
6DXQ
 
 | | Crystal structure of the LigJ Hydratase product complex with 4-carboxy-4-hydroxy-2-oxoadipate | | Descriptor: | (2S)-2-hydroxy-4-oxobutane-1,2,4-tricarboxylic acid, 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M. | | Deposit date: | 2018-06-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
6H3H
 
 | | Fab fragment of antibody against fullerene C60 | | Descriptor: | Anti-fullerene antibody Fab fragment Heavy chain, Anti-fullerene antibody Fab fragment Light chain, GLYCEROL, ... | | Authors: | Osipov, E.M, Tikhonova, T.V. | | Deposit date: | 2018-07-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Anti-C60 Fullerene Antibody Fab Fragment: Structural Determinants of Fullerene Binding.
Acta Naturae, 11, 2019
|
|
6PW8
 
 | |
2W3M
 
 | |
