7HSF
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z1503180352 | | Descriptor: | 6-methoxy-1,3,4,5-tetrahydro-2H-1-benzazepin-2-one, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5ERP
 
 | | Crystal structure of human Desmocollin-2 ectodomain fragment EC2-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2015-11-14 | | Release date: | 2016-06-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of adhesive binding by desmocollins and desmogleins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5QHP
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000554a | | Descriptor: | 1,2-ETHANEDIOL, 5-methoxy-2-(1~{H}-pyrazol-3-yl)phenol, Protein FAM83B | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5DF8
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH CEFOPERAZONE | | Descriptor: | (2R,4R,5R)-2-[(1R)-1-{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}-2-oxoethyl]-5-methyl-1,3-thiazinane-4-carboxylic acid, CHLORIDE ION, Cell division protein, ... | | Authors: | Ren, J, Nettleship, J.E, Males, A, Stuart, D.I, Owens, R.J. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms.
Febs Lett., 590, 2016
|
|
1ZZ2
 
 | | Two Classes of p38alpha MAP Kinase Inhibitors Having a Common Diphenylether Core but Exhibiting Divergent Binding Modes | | Descriptor: | Mitogen-activated protein kinase 14, N-[3-(4-FLUOROPHENOXY)PHENYL]-4-[(2-HYDROXYBENZYL)AMINO]PIPERIDINE-1-SULFONAMIDE, octyl beta-D-glucopyranoside | | Authors: | Michelotti, E.L, Moffett, K.K, Springman, E.B, Karpusas, M. | | Deposit date: | 2005-06-13 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two classes of p38alpha MAP kinase inhibitors having a common diphenylether core but exhibiting divergent binding modes.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7DNC
 
 | | Crystal structure of EV71 3C proteinase in complex with a novel inhibitor | | Descriptor: | 3C protease, ~{N}-[(2~{S})-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-05 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Peptidomimetic Aldehydes as Broad-Spectrum Inhibitors against Enterovirus and SARS-CoV-2.
J.Med.Chem., 65, 2022
|
|
3ILR
 
 | | Structure of Heparinase I from Bacteroides thetaiotaomicron in complex with tetrasaccharide product | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(4-1)-4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid, 4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ... | | Authors: | Garron, M.L, Cygler, M, Shaya, D. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I.
J.Biol.Chem., 284, 2009
|
|
5D0V
 
 | |
5QHO
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000010a | | Descriptor: | 1,2-ETHANEDIOL, Protein FAM83B, di(piperidin-1-yl)methanone | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1LKT
 
 | | CRYSTAL STRUCTURE OF THE HEAD-BINDING DOMAIN OF PHAGE P22 TAILSPIKE PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
J.Mol.Biol., 267, 1997
|
|
5QIX
 
 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0103007 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-phenylprop-2-yn-1-yl)acetamide, UNKNOWN LIGAND, ... | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
7E36
 
 | | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkanesulfonate monooxygenase SsuD/methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase (Luciferase family), ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis.
Nat Commun, 12, 2021
|
|
3IPS
 
 | | X-ray structure of benzisoxazole synthetic agonist bound to the LXR-alpha | | Descriptor: | Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, SULFATE ION, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
1N5U
 
 | | X-RAY STUDY OF HUMAN SERUM ALBUMIN COMPLEXED WITH HEME | | Descriptor: | MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SERUM ALBUMIN | | Authors: | Wardell, M, Wang, Z, Ho, J.X, Robert, J, Ruker, F, Ruble, J, Carter, D.C. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Atomic Structure of Human Methemalbumin at 1.9 A
Biochem.Biophys.Res.Commun., 291, 2002
|
|
5U94
 
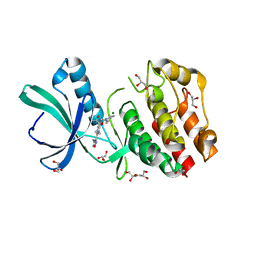 | | Crystal structure of the Mycobacterium tuberculosis PASTA kinase PknB in complex with the potential theraputic kinase inhibitor GSK690693. | | Descriptor: | 4-{2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-[(3S)-piperidin-3-ylmethoxy]-1H-imidazo[4,5-c]pyridin-4-yl}-2-methylbut-3 -yn-2-ol, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wlodarchak, N, Satyshur, K, Striker, R. | | Deposit date: | 2016-12-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Silico Screen and Structural Analysis Identifies Bacterial Kinase Inhibitors which Act with beta-Lactams To Inhibit Mycobacterial Growth.
Mol. Pharm., 15, 2018
|
|
2PUL
 
 | |
4LDQ
 
 | |
1VHH
 
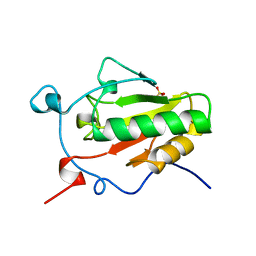 | | A POTENTIAL CATALYTIC SITE WITHIN THE AMINO-TERMINAL SIGNALLING DOMAIN OF SONIC HEDGEHOG | | Descriptor: | SONIC HEDGEHOG, SULFATE ION, ZINC ION | | Authors: | Hall, T.M.T, Porter, J.A, Beachy, P.A, Leahy, D.J. | | Deposit date: | 1995-10-03 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog.
Nature, 378, 1995
|
|
6I77
 
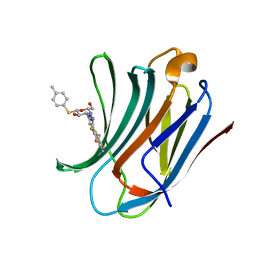 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative-4 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[4-azanyl-2,3,5,6-tetrakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
5CZ9
 
 | |
1QKE
 
 | | ERABUTOXIN | | Descriptor: | ERABUTOXIN A, SULFATE ION | | Authors: | Nastopoulos, V, Kanellopoulos, P.N, Tsernoglou, D. | | Deposit date: | 1998-01-16 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of dimeric and monomeric erabutoxin a refined at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1NJU
 
 | | Complex structure of HCMV Protease and a peptidomimetic inhibitor | | Descriptor: | Assemblin, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
6SU8
 
 | | Highly thermostable endoglucanase Cel7B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucanase, ... | | Authors: | Schiano-di-Cola, C, Morth, J.P, Westh, P, Borch, K. | | Deposit date: | 2019-09-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and biochemical characterization of a family 7 highly thermostable endoglucanase from the fungus Rasamsonia emersonii.
Febs J., 287, 2020
|
|
4HDR
 
 | | Crystal Structure of ArsAB in Complex with 5,6-dimethylbenzimidazole | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIMETHYLBENZIMIDAZOLE, ArsA, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4ZH1
 
 | | Complement factor H in complex with the GM1 glycan | | Descriptor: | Complement C3, Complement factor H, GLYCEROL, ... | | Authors: | Blaum, B.S, Stehle, T. | | Deposit date: | 2015-04-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Complement Factor H and Simian Virus 40 bind the GM1 ganglioside in distinct conformations.
Glycobiology, 26, 2016
|
|
