6C3Z
 
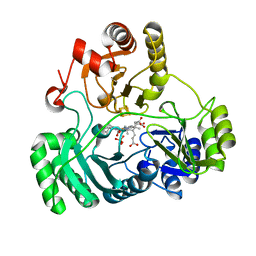 | | T477A SiRHP | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stroupe, M.E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | The role of extended Fe4S4cluster ligands in mediating sulfite reductase hemoprotein activity.
Biochim. Biophys. Acta, 1866, 2018
|
|
6D26
 
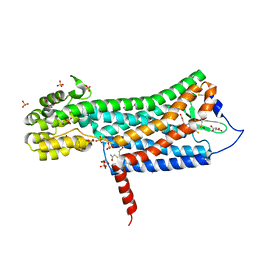 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant | | Descriptor: | OLEIC ACID, Prostaglandin D2 receptor 2, Endolysin chimera, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
1OVR
 
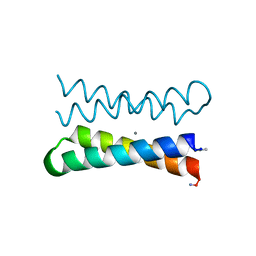 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL di-Mn(II)-DF1-L13 | | Descriptor: | MANGANESE (II) ION, four-helix bundle model di-Mn(II)-DF1-L13 | | Authors: | Di Costanzo, L, Geremia, S. | | Deposit date: | 2003-03-27 | | Release date: | 2004-05-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Response of a designed metalloprotein to changes in metal ion coordination, exogenous ligands, and active site volume determined by X-ray crystallography.
J.Am.Chem.Soc., 127, 2005
|
|
3ORV
 
 | |
4MBY
 
 | | Structure of B-Lymphotropic Polyomavirus VP1 in complex with 3'-sialyllactose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Khan, Z.M, Neu, U, Stehle, T. | | Deposit date: | 2013-08-21 | | Release date: | 2013-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structures of B-Lymphotropic Polyomavirus VP1 in Complex with Oligosaccharide Ligands.
Plos Pathog., 9, 2013
|
|
6D27
 
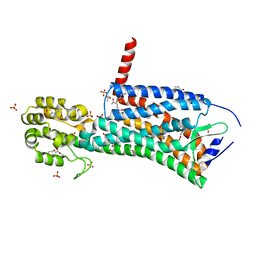 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
1K1O
 
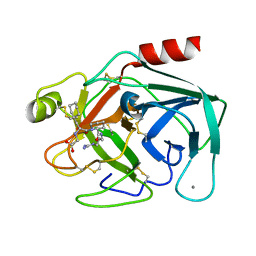 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, {[(1R)-2-((2S)-2-{[(3-{[AMINO(IMINO)METHYL]AMINO}PROPYL)AMINO]CARBONYL}PIPERIDINYL)-1-(CYCLOHEXYLMETHYL)-2-OXOETHYL]AMINO}ACETIC ACID | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
1K22
 
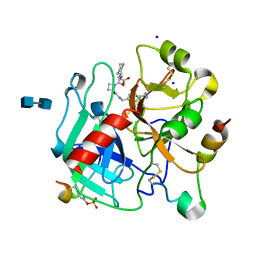 | | HUMAN THROMBIN-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-2, Prothrombin, ... | | Authors: | Stubbs, M.T, Musil, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
6CC0
 
 | |
1K1N
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, [N-[N-(4-METHOXY-2,3,6-TRIMETHYLPHENYLSULFONYL)-L-ASPARTYL]-D-(4-AMIDINO-PHENYLALANYL)]-PIPERIDINE | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
1K1I
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, N-ALPHA-(2-NAPHTHYLSULFONYL)-N-(3-AMIDINO-L-PHENYLALANINYL)-D-PIPECOLINIC ACID, TRYPSIN | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
1K1L
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, N-ALPHA-(2-NAPHTHYLSULFONYL)-3-AMIDINO-L-PHENYLALANINE PIPERAZIDE, SULFATE ION, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
4A7Q
 
 | | Structure of human I113T SOD1 mutant complexed with 4-(4-methyl-1,4- diazepan-1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYL-1,4-DIAZEPAN-1-YL)QUINAZOLINE, COPPER (II) ION, SULFATE ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-Ray Crystallography and Computational Docking for the Detection and Development of Protein-Ligand Interactions.
Curr.Med.Chem., 20, 2013
|
|
6H0U
 
 | | Glycogen synthase kinase-3 beta (GSK3) complex with a covalent [1,2,4]triazolo[1,5-a][1,3,5]triazine inhibitor | | Descriptor: | (2~{R})-3-[7-azanyl-5-(cyclohexylamino)-[1,2,4]triazolo[1,5-a][1,3,5]triazin-2-yl]-2-cyano-propanamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marcovich, I, Demitri, N, De Zorzi, R, Storici, P. | | Deposit date: | 2018-07-10 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Triazolotriazine-Based Dual GSK-3 beta /CK-1 delta Ligand as a Potential Neuroprotective Agent Presenting Two Different Mechanisms of Enzymatic Inhibition.
Chemmedchem, 14, 2019
|
|
4A7G
 
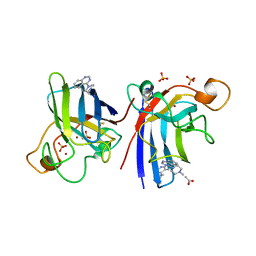 | | Structure of human I113T SOD1 mutant complexed with 4-methylpiperazin- 1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)QUINAZOLINE, ACETATE ION, COPPER (II) ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Sharma, R, Antonyuk, S.V, Strange, R.W, Berry, N.G, O'Neil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | X-ray crystallography and computational docking for the detection and development of protein-ligand interactions.
Curr.Med.Chem., 20, 2013
|
|
1K1P
 
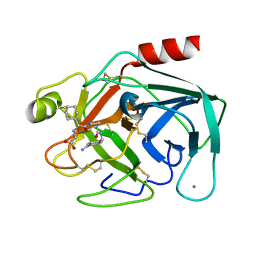 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, [((1R)-2-{(2S)-2-[({4-[AMINO(IMINO)METHYL]BENZYL}AMINO)CARBONYL]AZETIDINYL}-1-CYCLOHEXYL-2-OXOETHYL)AMINO]ACETIC ACID | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
1K1J
 
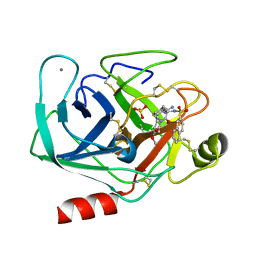 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, N-ALPHA-(2-NAPHTHYLSULFONYL)-N(3-AMIDINO-L-PHENYLALANINYL)ISOPIPECOLINIC ACID METHYL ESTER, SULFATE ION, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
1K1M
 
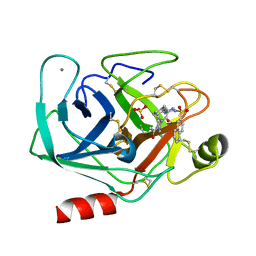 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, N-ALPHA-(2-NAPHTHYLSULFONYL)-N(3-AMIDINO-L-PHENYLALANINYL)-4-ACETYL-PIPERAZINE, SULFATE ION, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
3QA3
 
 | | Crystal Structure of A-domain in complex with antibody | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy chain, Antibody Light chain, ... | | Authors: | Mahalingam, B, Xiong, J.P, Arnaout, M.A. | | Deposit date: | 2011-01-10 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stable Coordination of the Inhibitory Ca2+ Ion at the Metal Ion-Dependent Adhesion Site in Integrin CD11b/CD18 by an Antibody-Derived Ligand Aspartate: Implications for Integrin Regulation and Structure-Based Drug Design.
J.Immunol., 187, 2011
|
|
6J0Q
 
 | | Crystal structure of P domain from GII.11 swine norovirus | | Descriptor: | VP1 capsid protein | | Authors: | Chen, Y. | | Deposit date: | 2018-12-25 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
3OUZ
 
 | | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, D-MALATE, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
1MQH
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Bromo-Willardiine at 1.8 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MQG
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
4JB6
 
 | | Structure of Pseudomonas aeruginosa FabF mutant C164Q | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, POTASSIUM ION | | Authors: | Baum, B, Lecker, L. | | Deposit date: | 2013-02-19 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Pseudomonas aeruginosa beta-ketoacyl-(acyl-carrier-protein) synthase II (FabF) and a C164Q mutant provide templates for antibacterial drug discovery and identify a buried potassium ion and a ligand-binding site that is an artefact of the crystal form.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
2AST
 
 | | Crystal structure of Skp1-Skp2-Cks1 in complex with a p27 peptide | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
