8QX6
 
 | | Novel laminarin-binding CBM X584 | | Descriptor: | PKD domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Zuehlke, M.K, Jeudy, A, Czjzek, M. | | Deposit date: | 2023-10-22 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ.Microbiol., 26, 2024
|
|
6D4P
 
 | | Ube2D1 in complex with ubiquitin variant Ubv.D1.1 | | Descriptor: | Ubiquitin Variant Ubv.D1.1, Ubiquitin-conjugating enzyme E2 D1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D5X
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase bound to ATP, Adenosylcobalamin, and Triphosphate | | Descriptor: | 5'-DEOXYADENOSINE, ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, ... | | Authors: | Dodge, G.J, Campanello, G, Smith, J.L, Banerjee, R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sacrificial Cobalt-Carbon Bond Homolysis in Coenzyme B12as a Cofactor Conservation Strategy.
J. Am. Chem. Soc., 140, 2018
|
|
4XUU
 
 | |
6Q8Z
 
 | | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide
To Be Published
|
|
6Q91
 
 | | Structure of human galactokinase 1 bound with 5-Chloro-N-isobutyl-2-methoxybenzamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 5-chloranyl-2-methoxy-~{N}-(2-methylpropyl)benzamide, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 5-Chloro-N-isobutyl-2-methoxybenzamide
To Be Published
|
|
5ID7
 
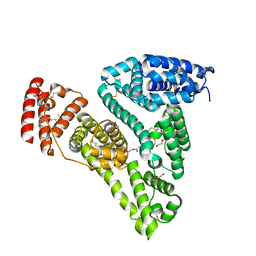 | | Crystal structure of human serum albumin in complex with phosphorodithioate derivative of myristoyl cyclic phosphatidic acid (cPA) | | Descriptor: | (4S)-2-sulfanylidene-4-[(tetradecanoyloxy)methyl]-1,3,2lambda~5~-dioxaphospholane-2-thiolate, DI(HYDROXYETHYL)ETHER, Serum albumin, ... | | Authors: | Sekula, B, Bujacz, A, Rytczak, P, Bujacz, G. | | Deposit date: | 2016-02-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural evidence of the species-dependent albumin binding of the modified cyclic phosphatidic acid with cytotoxic properties.
Biosci.Rep., 36, 2016
|
|
5UYK
 
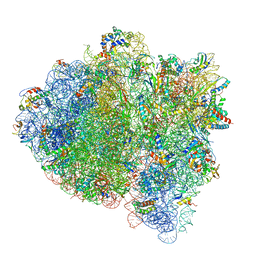 | | 70S ribosome bound with cognate ternary complex not base-paired to A site codon (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
5IDP
 
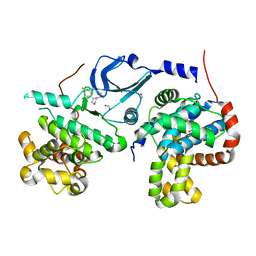 | | CDK8-CYCC IN COMPLEX WITH (3-Amino-1H-indazol-5-yl)-[(S)-2-(4-fluoro-phenyl)-piperidin-1-yl]-methanone | | Descriptor: | (3-amino-1H-indazol-5-yl)[(2S)-2-(4-fluorophenyl)piperidin-1-yl]methanone, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A, Czodrowski, P, Schiemann, K. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered by High-Throughput Screening.
J. Med. Chem., 59, 2016
|
|
7M4S
 
 | |
5UYD
 
 | |
5IDU
 
 | |
7LS2
 
 | | 80S ribosome from mouse bound to eEF2 (Class I) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
5IMR
 
 | | Structure of ribosome bound to cofactor at 5.7 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
5UYP
 
 | | 70S ribosome bound with near-cognate ternary complex base-paired to A site codon, open 30S (Structure II-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
7M0S
 
 | |
5IPW
 
 | | oligopeptide-binding protein OppA | | Descriptor: | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein, putative | | Authors: | Lee, H.H, Kim, H.J, Yoon, H.J. | | Deposit date: | 2016-03-10 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a putative oligopeptide-binding periplasmic protein from a hyperthermophile
Extremophiles, 20, 2016
|
|
5IPY
 
 | | Crystal structure of WT RnTmm | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
7LTN
 
 | | Crystal structure of Mpro in complex with inhibitor CDD-1713 | | Descriptor: | 2-[4-(1~{H}-indazol-4-yl)-2-methanoyl-6-methoxy-phenoxy]-~{N},~{N}-dimethyl-ethanamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M, Young, D, Melek, N, Chamakuri, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 M pro inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5V3I
 
 | |
5IQC
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia (CTD of AAC(6')-Ie/APH(2'')-Ia) in complex with GMPPNP, Magnesium, and Gentamicin C1 | | Descriptor: | Bifunctional AAC/APH, CHLORIDE ION, GLYCEROL, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antibiotic Binding Drives Catalytic Activation of Aminoglycoside Kinase APH(2)-Ia.
Structure, 24, 2016
|
|
7M7E
 
 | | 6-Deoxyerythronolide B synthase (DEBS) hybrid module (M3/1) in complex with antibody fragment 1B2 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), 6-deoxyerythronolide-B synthase EryA2, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
5IQN
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitution Q134E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_SRIRIRGYVR | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Protein FimF, ... | | Authors: | Giese, C, Eras, J, Kern, A, Scharer, M.A, Capitani, G, Glockshuber, R. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7LLI
 
 | | Stimulatory immune receptor protein complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Rice, M.T, Littler, D.R, Rossjohn, J, Gully, B.S. | | Deposit date: | 2021-02-04 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the antigen-presenting molecule MR1 by a V delta 3 + gamma delta T cell receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5V44
 
 | |
