6BIQ
 
 | |
7BIG
 
 | | Crystal structure of v13WRAP-T, a 7-bladed designer protein | | Descriptor: | CHLORIDE ION, v13WRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIE
 
 | | Crystal structure of nvWrap-T, a 7-bladed symmetric propeller | | Descriptor: | CITRIC ACID, nvWRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIF
 
 | | Crystal structure of v22WRAP-T, a 7-bladed designer protein | | Descriptor: | v22WRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
6TEI
 
 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the 2-aminothiazole-type inhibitor 17 | | Descriptor: | 3-[(4-pyridin-2-yl-1,3-thiazol-2-yl)amino]benzoic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
6CGY
 
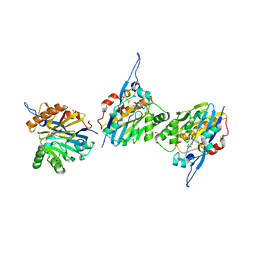 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to a phosphate anion | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
5M4R
 
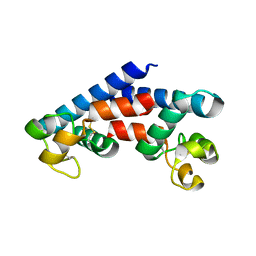 | | Structural tuning of CD81LEL (space group C2) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, SULFATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-19 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
6RHE
 
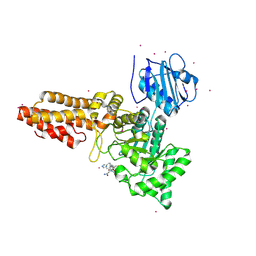 | | CpOGA D298N in complex with hOGA-derived S-GlcNAc peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-ALA-HIS-CYS-GLY-NH2, CADMIUM ION, ... | | Authors: | Van Aalten, D, Bartual, S.G, Gorelik, A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-12-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Genetic recoding to dissect the roles of site-specific protein O-GlcNAcylation.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2KDI
 
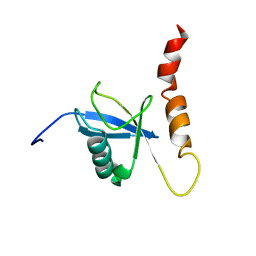 | | Solution structure of a Ubiquitin/UIM fusion protein | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein 27 fusion protein | | Authors: | Sgourakis, N.G, Patel, M.M, Garcia, A.E, Makhatadze, G.I, McCallum, S.A. | | Deposit date: | 2009-01-09 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Conformational Dynamics and Structural Plasticity Play Critical Roles in the Ubiquitin Recognition of a UIM Domain.
J.Mol.Biol., 396, 2010
|
|
5T5M
 
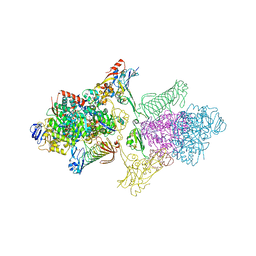 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, TRIGONAL FORM AT 2.5 A. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, GLYCEROL, HYDROSULFURIC ACID, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
6RZT
 
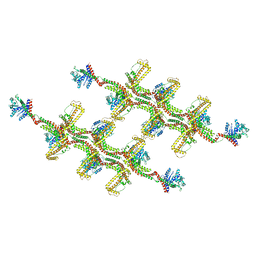 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
5JPX
 
 | |
5K1I
 
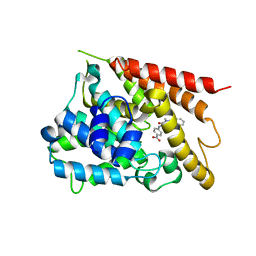 | | PDE4 crystal structure in complex with small molecule inhibitor | | Descriptor: | 4-[(5-acetyl-2-ethyl-3-oxo-6-phenyl-2,3-dihydropyridazin-4-yl)amino]benzoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Segarra, V, Hernandez, B, Ferrer-Miralles, N, Korndoerfer, I, Aymami, J. | | Deposit date: | 2016-05-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Biphenyl Pyridazinone Derivatives as Inhaled PDE4 Inhibitors: Structural Biology and Structure-Activity Relationships.
J. Med. Chem., 59, 2016
|
|
1AGQ
 
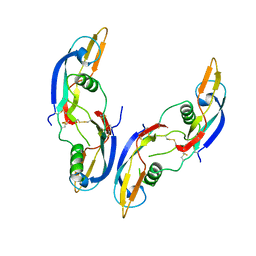 | | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR FROM RAT | | Descriptor: | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR | | Authors: | Eigenbrot, C, Gerber, N. | | Deposit date: | 1997-03-25 | | Release date: | 1997-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of glial cell-derived neurotrophic factor at 1.9 A resolution and implications for receptor binding.
Nat.Struct.Biol., 4, 1997
|
|
5KAJ
 
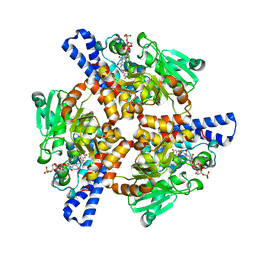 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21, A319C mutant | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[(~{E})-2-[3,5-bis(oxidanyl)phenyl]-1-oxidanyl-ethenyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3J6C
 
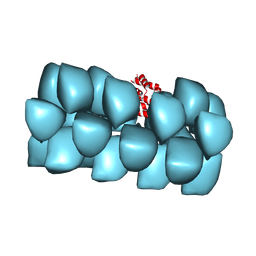 | | Cryo-EM structure of MAVS CARD filament | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Xu, H, He, X, Zheng, H, Huang, L.J, Hou, F, Yu, Z, de la Cruz, M.J, Borkowski, B, Zhang, X, Chen, Z.J, Jiang, Q.-X. | | Deposit date: | 2014-02-04 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Structural basis for the prion-like MAVS filaments in antiviral innate immunity.
Elife, 3, 2014
|
|
6TGU
 
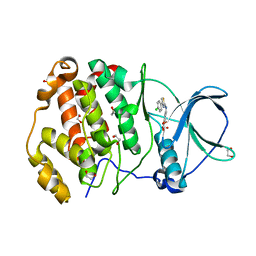 | | Crystal structure of human protein kinase CK2alpha'(CSNK2A2 gene product) in complex with the 2-aminothiazole-type inhibitor Cl-OH-3 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | Deposit date: | 2019-11-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.833 Å) | | Cite: | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
5GRK
 
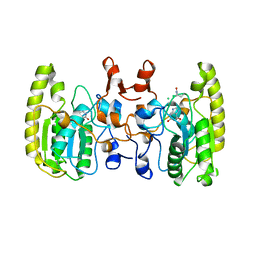 | | Crystal structure of Uracil DNA glycosylase -Xanthine complex from Bradyrhizobium diazoefficiens | | Descriptor: | Blr0248 protein, XANTHINE | | Authors: | Patil, V.V, Ullas, V.C, Ahn, W, Varshney, U, Woo, E. | | Deposit date: | 2016-08-11 | | Release date: | 2017-05-03 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Uracil DNA glycosylase (UDG) activities in Bradyrhizobium diazoefficiens: characterization of a new class of UDG with broad substrate specificity
Nucleic Acids Res., 45, 2017
|
|
8JKZ
 
 | |
6TGX
 
 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with a bisubstrate analogue | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA | | Authors: | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
8JL0
 
 | | Cryo-EM structure of the prokaryotic SPARSA system complex | | Descriptor: | DNA (5'-D(P*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain protein, ... | | Authors: | Xu, X, Zhen, X, Long, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antiphage immunity generated by a prokaryotic Argonaute-associated SPARSA system.
Nat Commun, 15, 2024
|
|
6CC4
 
 | | Structure of MurJ from Escherichia coli | | Descriptor: | PHOSPHATE ION, soluble cytochrome b562, lipid II flippase MurJ chimera | | Authors: | Zheng, S, Kruse, A.C. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mutagenic analysis of the lipid II flippase MurJ fromEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8AG1
 
 | | Crystal structure of a novel OX40 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Novel OX40 antibody heavy chain, Novel OX40 antibody light chain, ... | | Authors: | Gao, H, Zhou, A. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structural Basis of a Novel Agonistic Anti-OX40 Antibody.
Biomolecules, 12, 2022
|
|
1A5Z
 
 | | LACTATE DEHYDROGENASE FROM THERMOTOGA MARITIMA (TMLDH) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CADMIUM ION, L-LACTATE DEHYDROGENASE, ... | | Authors: | Auerbach, G, Ostendorp, R, Prade, L, Korndoerfer, I, Dams, T, Huber, R, Jaenicke, R. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization.
Structure, 6, 1998
|
|
8J8K
 
 | | Membrane bound PRTase, C3 symmetry, acceptor bound | | Descriptor: | Decaprenyl-phosphate phosphoribosyltransferase, MONO-TRANS, OCTA-CIS DECAPRENYL-PHOSPHATE | | Authors: | Wu, F.Y, Gao, S, Zhang, L, Rao, Z.H. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural analysis of phosphoribosyltransferase-mediated cell wall precursor synthesis in Mycobacterium tuberculosis.
Nat Microbiol, 9, 2024
|
|
