5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
1QFW
 
 | | TERNARY COMPLEX OF HUMAN CHORIONIC GONADOTROPIN WITH FV ANTI ALPHA SUBUNIT AND FV ANTI BETA SUBUNIT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY (ANTI ALPHA SUBUNIT) (HEAVY CHAIN), ANTIBODY (ANTI ALPHA SUBUNIT) (LIGHT CHAIN), ... | | Authors: | Tegoni, M, Spinelli, S, Cambillau, C. | | Deposit date: | 1999-04-15 | | Release date: | 2000-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of a ternary complex between human chorionic gonadotropin (hCG) and two Fv fragments specific for the alpha and beta-subunits.
J.Mol.Biol., 289, 1999
|
|
3ED4
 
 | | Crystal structure of putative arylsulfatase from escherichia coli | | Descriptor: | ARYLSULFATASE, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Gilmore, M, Chang, S, Bain, K, Wasserman, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Arylsulfatase from Escherichia Coli
To be Published
|
|
3EIS
 
 | |
5ZCI
 
 | |
5ZCM
 
 | |
8ZNJ
 
 | | Cryo-EM structure of a short prokaryotic Argonaute system from archaeon Suldolobus islandicus | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*CP*AP*GP*GP*GP*TP*AP*TP*CP*TP*AP*AP*GP*CP*TP*TP*TP*GP*AP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Dai, Z.K, Guan, Z.Y, Han, W.Y, Zou, T.T. | | Deposit date: | 2024-05-27 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic insights into the activation of a short prokaryotic argonaute system from archaeon Sulfolobus islandicus.
Nucleic Acids Res., 53, 2025
|
|
1FYP
 
 | |
6A9A
 
 | | Ternary complex crystal structure of dCH with dCMP and THF | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Deoxycytidylate 5-hydroxymethyltransferase, ... | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2018-07-12 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A cytosine modification mechanism revealed by the structure of a ternary complex of deoxycytidylate hydroxymethylase from bacteriophage T4 with its cofactor and substrate.
Iucrj, 6, 2019
|
|
7KC8
 
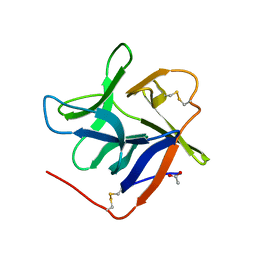 | | Salivary protein from Culex quinquefasciatus that belongs to the Cysteins and Tryptophan-Rich (CWRC) family | | Descriptor: | 16.4 kDa salivary peptide, ACETIC ACID | | Authors: | Garboczi, D.N, Gittis, A.G, Kern, O, Martin-Martin, I. | | Deposit date: | 2020-10-05 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structures of two salivary proteins from the West Nile vector Culex quinquefasciatus reveal a beta-trefoil fold with putative sugar binding properties
Curr Res Struct Biol, 3, 2021
|
|
9BGY
 
 | |
9BH0
 
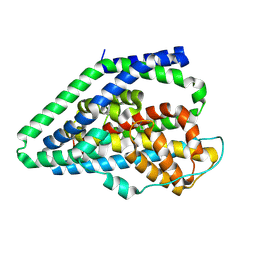 | |
9BH1
 
 | |
9BH2
 
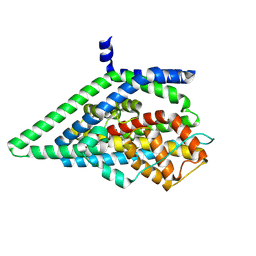 | | Apo GltPh, outward-facing state | | Descriptor: | Glutamate transporter homolog | | Authors: | Reddy, K.D, Boudker, O. | | Deposit date: | 2024-04-19 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Evolutionary analysis reveals the origin of sodium coupling in glutamate transporters.
Biorxiv, 2024
|
|
9BGZ
 
 | |
1O94
 
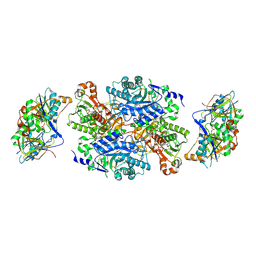 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O95
 
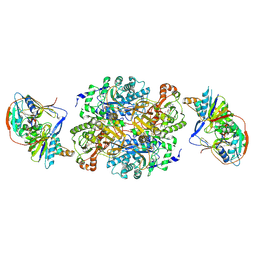 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1T02
 
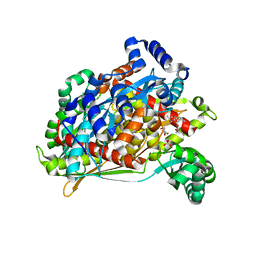 | | Crystal structure of a Statin bound to class II HMG-CoA reductase | | Descriptor: | (3R,5R)-7-((1R,2R,6S,8R,8AS)-2,6-DIMETHYL-8-{[(2R)-2-METHYLBUTANOYL]OXY}-1,2,6,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL)-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Tabernero, L, Rodwell, V.W, Stauffacher, C. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a statin bound to a class II hydroxymethylglutaryl-CoA reductase.
J.Biol.Chem., 278, 2003
|
|
1FYO
 
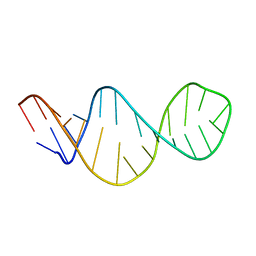 | |
2DNG
 
 | | Solution structure of RNA binding domain in Eukaryotic translation initiation factor 4H | | Descriptor: | Eukaryotic translation initiation factor 4H | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in Eukaryotic translation initiation factor 4H
To be Published
|
|
8X1X
 
 | | SARS-CoV-2 Papain like protease (PLpro) in complex with inhibitor Lithocholic acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, GLYCEROL, Papain-like protease nsp3, ... | | Authors: | Choudhary, S, Verma, S, Kumar, P, Tomar, S. | | Deposit date: | 2023-11-09 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SARS-CoV-2 Papain like protease (PLpro) in complex with inhibitor Lithocholic acid
To Be Published
|
|
6CDE
 
 | | Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDI
 
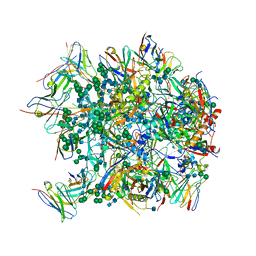 | | Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6AEF
 
 | | PapA2 acyl transferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Trehalose-2-sulfate acyltransferase PapA2, ... | | Authors: | Chaudhary, S, Rao, V, Panchal, V. | | Deposit date: | 2018-08-04 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A novel mutation alters the stability of PapA2 resulting in the complete abrogation of sulfolipids in clinical mycobacterial strains.
Faseb Bioadv, 1, 2019
|
|
3U7X
 
 | | Crystal structure of the human eIF4E-4EBP1 peptide complex without cap | | Descriptor: | Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1, SULFATE ION, ... | | Authors: | Siddiqui, N, Tempel, W, Nedyalkova, L, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Borden, K.L.B, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-14 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Allosteric Effects of 4EBP1 on the Eukaryotic Translation Initiation Factor eIF4E.
J.Mol.Biol., 415, 2012
|
|
