5K0M
 
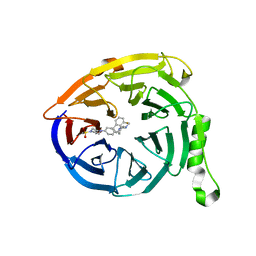 | | Targeting the PRC2 complex through a novel protein-protein interaction inhibitor of EED | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-{4-[4-(methylsulfonyl)piperazin-1-yl]phenyl}pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Bigelow, L.J, Zhu, H, Sun, C. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex.
Nat. Chem. Biol., 13, 2017
|
|
7T4P
 
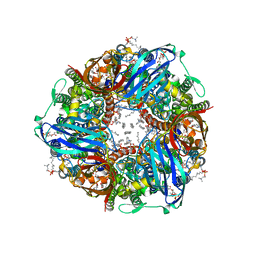 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO treated with potassium cyanide and copper in a native lipid nanodisc at 3.62 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7T4O
 
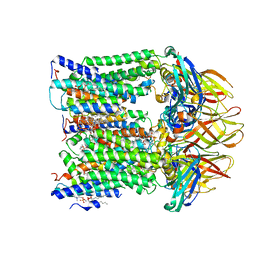 | |
5K1B
 
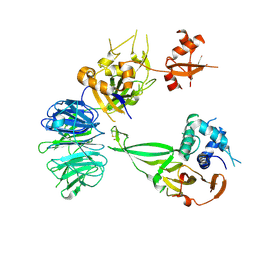 | |
5KDO
 
 | | Heterotrimeric complex of the 4 alanine insertion variant of the Gi alpha1 subunit and the Gbeta1-Ggamma1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Kaya, A.I, Lokits, A.D, Gilbert, J, Iverson, T.M, Meiler, J, Hamm, H.E. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Conserved Hydrophobic Core in G alpha i1 Regulates G Protein Activation and Release from Activated Receptor.
J.Biol.Chem., 291, 2016
|
|
5CR2
 
 | |
5CVL
 
 | | WDR48 (UAF-1), residues 2-580 | | Descriptor: | GOLD ION, PHOSPHATE ION, WD repeat-containing protein 48 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
6XAJ
 
 | | Crystal structure of NzeB | | Descriptor: | 1,2-ETHANEDIOL, NzeB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
5CWW
 
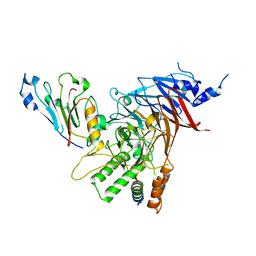 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5KHR
 
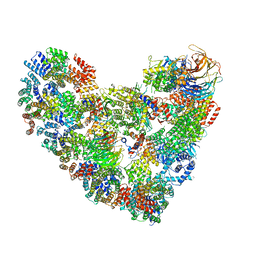 | | Model of human Anaphase-promoting complex/Cyclosome complex (APC15 deletion mutant) in complex with the E2 UBE2C/UBCH10 poised for ubiquitin ligation to substrate (APC/C-CDC20-substrate-UBE2C) | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | VanderLinden, R, Yamaguchi, M, Dube, P, Haselbach, D, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation.
Mol.Cell, 63, 2016
|
|
4OKU
 
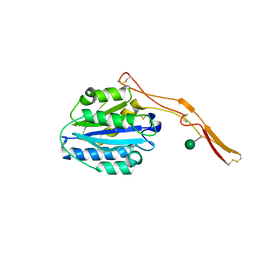 | | Structure of Toxoplasma gondii proMIC2 | | Descriptor: | Micronemal protein MIC2, alpha-D-mannopyranose | | Authors: | Song, G, Springer, T.A. | | Deposit date: | 2014-01-22 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the Toxoplasma gliding motility adhesin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OWR
 
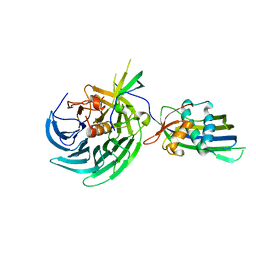 | | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair Rae1-Nup98 | | Descriptor: | Matrix protein, Nuclear pore complex protein Nup98-Nup96, mRNA export factor | | Authors: | Ren, Y, Quan, B, Seo, H.S, Blobel, G. | | Deposit date: | 2014-02-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 Nup98).
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6YBT
 
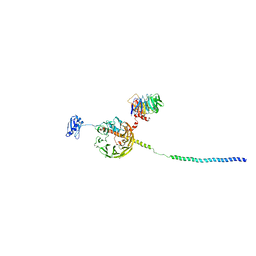 | | Structure of a human 48S translational initiation complex - eIF3bgi | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6Y7N
 
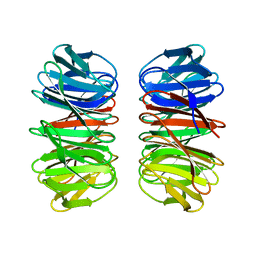 | | The crystal structure of the eight-bladed symmetrical designer protein Tako8 in the presence of tellurotungstic Anderson-Evans (TEW) | | Descriptor: | Tako8 | | Authors: | Vandebroek, L, Noguchi, H, Parac-Vogt, T.N, Van Meervelt, L, Voet, A.R.D. | | Deposit date: | 2020-03-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Shape and Size Complementarity-Induced Formation of Supramolecular Protein Assemblies with Metal-Oxo Clusters
Cryst.Growth Des., 2021
|
|
6Y7P
 
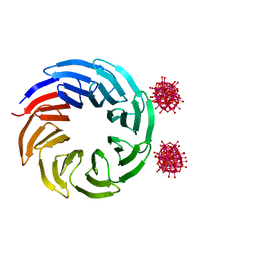 | | The complex between the eight-bladed symmetrical designer protein Tako8 and 1:2 zirconium(IV) Wells-Dawson (ZrWD) | | Descriptor: | Tako8, W-Zr-cluster | | Authors: | Vandebroek, L, Noguchi, H, Parac-Vogt, T.N, Van Meervelt, L, Voet, A.R.D. | | Deposit date: | 2020-03-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Shape and Size Complementarity-Induced Formation of Supramolecular Protein Assemblies with Metal-Oxo Clusters
Cryst.Growth Des., 2021
|
|
5L8W
 
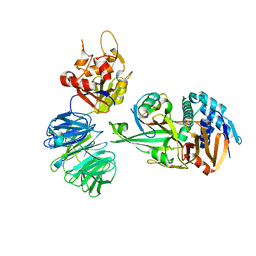 | | Structure of USP12-UB-PRG/UAF1 | | Descriptor: | GLYCEROL, Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Dharadhar, S, Sixma, T. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme.
J.Struct.Biol., 196, 2016
|
|
6YLH
 
 | |
5KQQ
 
 | |
6YLF
 
 | |
5EAP
 
 | | Crystal structure of human WDR5 in complex with compound 9e | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
6YBS
 
 | | Structure of a human 48S translational initiation complex - head | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
5KQN
 
 | |
6Y7O
 
 | | The complex between the eight-bladed symmetrical designer protein Tako8 and the silicotungstic acid Keggin (STA) | | Descriptor: | Keggin (STA), Tako8 | | Authors: | Vandebroek, L, Noguchi, H, Parac-Vogt, T.N, Van Meervelt, L, Voet, A.R.D. | | Deposit date: | 2020-03-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Shape and Size Complementarity-Induced Formation of Supramolecular Protein Assemblies with Metal-Oxo Clusters
Cryst.Growth Des., 2021
|
|
6YAL
 
 | | Mammalian 48S late-stage initiation complex with beta-globin mRNA | | Descriptor: | 18S ribosomal RNA, 40S Ribosomal protein uS3, 40S Ribosomal protein uS7, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
8I9P
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Mak16 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, 60S ribosomal protein L16-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
