2WV8
 
 | | Complex of human dihydroorotate dehydrogenase with the inhibitor 221290 | | Descriptor: | 2-ACETAMIDO-5-(4-PHENYLPHENYL)BENZOIC ACID, ACETATE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Walse, B, Svensson, B, Fritzson, I, Dahlberg, L, Wellmar, U, Al-Karadaghi, S. | | Deposit date: | 2009-10-15 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of Human Dhodh by 4-Hydroxycoumarins, Fenamic Acids, and N-(Alkylcarbonyl)Anthranilic Acids Identified by Structure-Guided Fragment Selection.
Chemmedchem, 5, 2010
|
|
2VZ1
 
 | | Premat-galactose oxidase | | Descriptor: | ACETATE ION, CALCIUM ION, GALACTOSE OXIDASE | | Authors: | Rogers, M.S, Hurtado-Guerrero, R, Firbank, S.J, Halcrow, M.A, Dooley, D.M, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cross-Link Formation of the Cysteine 228-Tyrosine 272 Catalytic Cofactor of Galactose Oxidase Does not Require Dioxygen.
Biochemistry, 47, 2008
|
|
8D9P
 
 | | De Novo Photosynthetic Reaction Center Protein Equipped with Heme B and Mn(II) cations | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of photosynthetic reaction center protein maquettes.
Front Mol Biosci, 9, 2022
|
|
6K26
 
 | | Crystal structure of Vibrio cholerae methionine aminopeptidase | | Descriptor: | Methionine aminopeptidase, SODIUM ION | | Authors: | Pillalamarri, V, Addlagatta, A. | | Deposit date: | 2019-05-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Methionine aminopeptidases with short sequence inserts within the catalytic domain are differentially inhibited: Structural and biochemical studies of three proteins from Vibrio spp.
Eur.J.Med.Chem., 209, 2020
|
|
9JGC
 
 | | Crystal structure of Nep1 in complex with adenosine from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, CHLORIDE ION, ... | | Authors: | Saha, S, Kanaujia, S.P. | | Deposit date: | 2024-09-06 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural analysis of the ribosome assembly factor Nep1, an N1-specific pseudouridine methyltransferase, reveals mechanistic insights.
Febs J., 292, 2025
|
|
4F9M
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | (9aS)-8-acetyl-N-[(2-ethyl-4-fluoronaphthalen-1-yl)methyl]-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peroxisome proliferator-activated receptor gamma, peptide from Nuclear receptor coactivator 1 | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2012-05-19 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and biological evaluation of novel (-)-cercosporamide derivatives as potent selective PPARg modulators
Eur.J.Med.Chem., 54, 2012
|
|
9JGB
 
 | | Crystal structure of Nep1 from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ribosomal RNA small subunit methyltransferase Nep1, ... | | Authors: | Saha, S, Kanaujia, S.P. | | Deposit date: | 2024-09-06 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of the ribosome assembly factor Nep1, an N1-specific pseudouridine methyltransferase, reveals mechanistic insights.
Febs J., 292, 2025
|
|
9JGD
 
 | | Crystal structure of Nep1 in complex with 5'-methylthioadenosine from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ACETATE ION, ... | | Authors: | Saha, S, Kanaujia, S.P. | | Deposit date: | 2024-09-06 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the ribosome assembly factor Nep1, an N1-specific pseudouridine methyltransferase, reveals mechanistic insights.
Febs J., 292, 2025
|
|
2VZ3
 
 | | bleached galactose oxidase | | Descriptor: | ACETATE ION, COPPER (II) ION, GALACTOSE OXIDASE | | Authors: | Rogers, M.S, Hurtado-Guerrero, R, Firbank, S.J, Halcrow, M.A, Dooley, D.M, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Link Formation of the Cysteine 228-Tyrosine 272 Catalytic Cofactor of Galactose Oxidase Does not Require Dioxygen.
Biochemistry, 47, 2008
|
|
2XMH
 
 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CITRATE ANION, CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Archaeoglobus fulgidus CTP:inositol-1-phosphate cytidylyltransferase, a key enzyme for di-myo-inositol-phosphate synthesis in (hyper)thermophiles.
J. Bacteriol., 193, 2011
|
|
4HEE
 
 | | Crystal structure of PPARgamma in complex with compound 13 | | Descriptor: | 5-benzyl-2-ethyl-3-{(1S)-5-[2-(1H-tetrazol-5-yl)phenyl]-2,3-dihydro-1H-inden-1-yl}-3,5-dihydro-4H-imidazo[4,5-c]pyridin-4-one, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Han, S. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and evaluation of imidazo[4,5-c]pyridin-4-one derivatives with dual activity at angiotensin II type 1 receptor and peroxisome proliferator-activated receptor-gamma
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4D8W
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase soaked with D-cys shows pyruvate bound 4 A away from active site | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4CMG
 
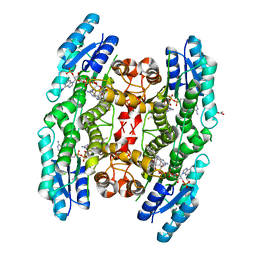 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 6-(4-fluorophenyl)-5-(4-methoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
5MTY
 
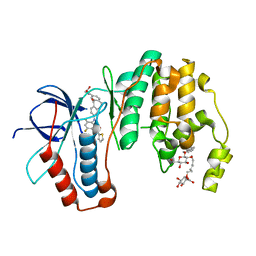 | | Dibenzosuberone inhibitor 8e in complex with p38 MAPK | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside, ~{N}-[2,4-bis(fluoranyl)-5-[[14-(2-hydroxyethylcarbamoyl)-2-oxidanylidene-6-tricyclo[9.4.0.0^{3,8}]pentadeca-1(15),3(8),4,6,11,13-hexaenyl]amino]phenyl]thiophene-2-carboxamide | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-01-11 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Novel Type I(1)/2 p38 alpha MAP Kinase Inhibitors with Excellent Selectivity, High Potency, and Prolonged Target Residence Time by Interfering with the R-Spine.
J. Med. Chem., 60, 2017
|
|
4CMC
 
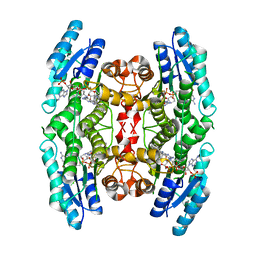 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | N4-cyclohexyl-5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 1 | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CM9
 
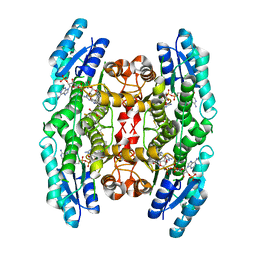 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-amino-5,6-diphenyl-3H-pyrrolo[2,3-d]pyrimidin-4(7H)-one, ACETATE ION, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CLH
 
 | |
2VKI
 
 | | Structure of the PDK1 PH domain K465E mutant | | Descriptor: | 3-PHOPSHOINOSITIDE DEPENDENT PROTEIN KINASE 1, GLYCEROL, SULFATE ION | | Authors: | Komander, D, Bayascas, J.R, Deak, M, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutation of the Pdk1 Ph Domain Inhibits Protein Kinase B/Akt, Leading to Small Size and Insulin Resistance.
Mol.Cell.Biol., 28, 2008
|
|
4CME
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | N4,N4-dimethyl-5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 1 | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CLD
 
 | |
4CMJ
 
 | |
2V79
 
 | | Crystal Structure of the N-terminal domain of DnaD from Bacillus Subtilis | | Descriptor: | CHLORIDE ION, DNA REPLICATION PROTEIN DNAD, SODIUM ION | | Authors: | Schneider, S, Zhang, W, Soultanas, P, Paoli, M. | | Deposit date: | 2007-07-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-Terminal Oligomerization Domain of Dnad Reveals a Unique Tetramerization Motif and Provides Insights Into Scaffold Formation.
J.Mol.Biol., 376, 2008
|
|
5N65
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9h | | Descriptor: | 2-phenyl-~{N}4-(2-thiophen-2-ylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
4CM8
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | (E)-2,4-diamino-6-(4-methylstyryl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
2VNG
 
 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens in complex with blood group A-trisaccharide ligand. | | Descriptor: | CALCIUM ION, CPE0329, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
