4LGP
 
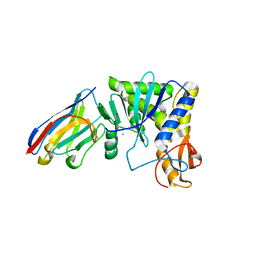 | | Ricin A chain bound to camelid nanobody (VHH1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ricin, ... | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M, Burshteyn, F, Cassidy, M, Gary, E, Mantis, N. | | Deposit date: | 2013-06-28 | | Release date: | 2014-06-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Ricin Toxin's Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies.
J.Mol.Biol., 426, 2014
|
|
4LHQ
 
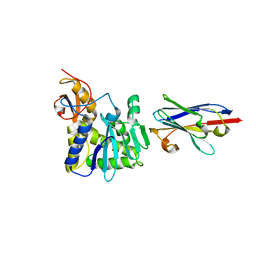 | | Ricin A chain bound to camelid nanobody (VHH8) | | Descriptor: | Camelid nanobody, Ricin | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M, Burshteyn, F, Cassidy, M, Gary, E, Mantis, N. | | Deposit date: | 2013-07-01 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ricin Toxin's Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies.
J.Mol.Biol., 426, 2014
|
|
5WQS
 
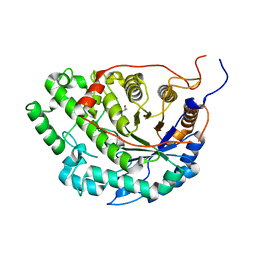 | | Crystal structure of Apo Beta-Amylase from Sweet potato | | Descriptor: | Beta-amylase, ISOPROPYL ALCOHOL | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
5WDR
 
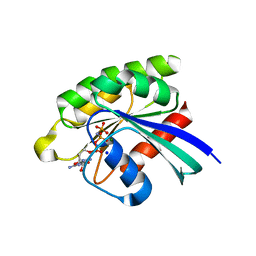 | | Choanoflagellate Salpingoeca rosetta Ras with GMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras protein, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
4J2R
 
 | | Middle domain of influenza A virus RNA-dependent polymerase PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Qiu, H, Tsurumura, T, Tsumori, Y, Hatakeyama, D, Kuzuhara, T, Tsuge, H. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystallization and preliminary X-ray diffraction studies of a surface mutant of the middle domain of PB2 from human influenza A (H1N1) virus
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
5A8O
 
 | | Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with cellotetraose | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
3ITF
 
 | |
3KDZ
 
 | |
1HQM
 
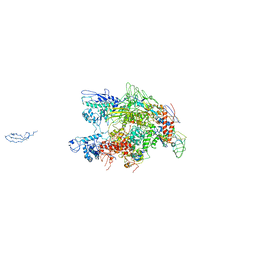 | | CRYSTAL STRUCTURE OF THERMUS AQUATICUS CORE RNA POLYMERASE-INCLUDES COMPLETE STRUCTURE WITH SIDE-CHAINS (EXCEPT FOR DISORDERED REGIONS)-FURTHER REFINED FROM ORIGINAL DEPOSITION-CONTAINS ADDITIONAL SEQUENCE INFORMATION | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Minakhin, L, Bhagat, S, Brunning, A, Campbell, E.A, Darst, S.A, Ebright, R.H, Severinov, K. | | Deposit date: | 2000-12-18 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4M7B
 
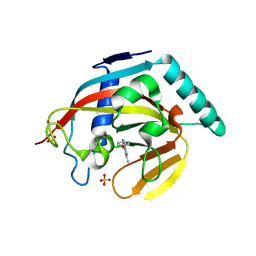 | | Human tankyrase 2 - catalytic Parp domain in complex with an inhibitor UPF1854 | | Descriptor: | 4-{2-[(6-methyl[1,2,4]triazolo[4,3-b]pyridazin-8-yl)amino]ethyl}phenol, SULFATE ION, Tankyrase-2, ... | | Authors: | Karlberg, T, Camaioni, E, Schuler, H. | | Deposit date: | 2013-08-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, Crystallographic Studies, and Preliminary Biological Appraisal of New Substituted Triazolo[4,3-b]pyridazin-8-amine Derivatives as Tankyrase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4M92
 
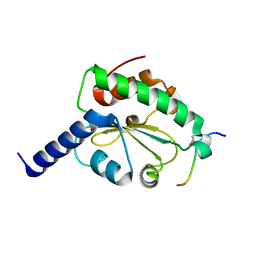 | | Crystal structure of hN33/Tusc3-peptide 2 | | Descriptor: | Interleukin-1 receptor accessory protein-like 1, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4MHE
 
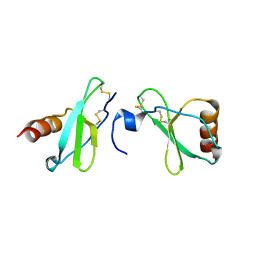 | | Crystal structure of CC-chemokine 18 | | Descriptor: | ACETATE ION, C-C motif chemokine 18 | | Authors: | Liang, W.G, Tang, W.-J. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
5AMB
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with amyloid-beta 35-42 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMYLOID BETA A4 PROTEIN, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Masuyer, G, Larmuth, K.M, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Kinetic and Structural Characterisation of Amyloid-Beta Metabolism by Human Angiotensin-1- Converting Enzyme (Ace)
FEBS J., 283, 2016
|
|
5ARL
 
 | |
5AMC
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with amyloid-beta fluorogenic fragment 4-10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Masuyer, G, Larmuth, K.M, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Kinetic and Structural Characterisation of Amyloid-Beta Metabolism by Human Angiotensin-1- Converting Enzyme (Ace)
FEBS J., 283, 2016
|
|
6MGT
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase Mutant H110A | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Yang, Y, Daivs, I, Matsui, T, Rubalcava, I, Liu, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
5BNF
 
 | | Apo structure of porcine CD38 | | Descriptor: | Uncharacterized protein | | Authors: | Ting, K.Y, Leung, C.P.F, Graeff, R.M, Lee, H.C, Hao, Q, Kotaka, M. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Porcine CD38 exhibits prominent secondary NAD(+) cyclase activity.
Protein Sci., 25, 2016
|
|
3N7O
 
 | | X-ray structure of human chymase in complex with small molecule inhibitor. | | Descriptor: | (S)-[(1S)-1-(5-chloro-1-benzothiophen-3-yl)-2-{[(E)-2-(3,4-difluorophenyl)ethenyl]amino}-2-oxoethyl]methylphosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase, ... | | Authors: | Abad, M.C, Kervinen, J, Crysler, C, Bayoumy, S, Spurlino, J, Deckman, I, Greco, M.N, Maryanoff, B.E, Degaravilla, L. | | Deposit date: | 2010-05-27 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potency variation of small-molecule chymase inhibitors across species.
Biochem. Pharmacol., 80, 2010
|
|
5ARJ
 
 | |
6IF9
 
 | |
2JWM
 
 | |
6IZW
 
 | | Myxococcus xanthus MglA bound to GTP-gamma-S and MglB | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Gliding motility protein MglB, MAGNESIUM ION, ... | | Authors: | Baranwal, J, Gayathri, P. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric regulation of a prokaryotic small Ras-like GTPase contributes to cell polarity oscillations in bacterial motility.
Plos Biol., 17, 2019
|
|
5AR6
 
 | | crystal structure of porcine RNase 4 | | Descriptor: | 1,2-ETHANEDIOL, RIBONUCLEASE 4 | | Authors: | Liang, S, Acharya, K.R. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of Substrate Specificity in Porcine Rnase 4.
FEBS J., 283, 2016
|
|
6IXM
 
 | | Crystal structure of the ketone reductase ChKRED20 from the genome of Chryseobacterium sp. CA49 complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase reductase | | Authors: | Zhao, F.J, Jin, Y, Liu, Z.C, Wang, G.G, Wu, Z.L. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structure-guided engineering of ChKRED20 from Chryseobacterium sp. CA49 for asymmetric reduction of aryl ketoesters.
Enzyme Microb. Technol., 125, 2019
|
|
6IWI
 
 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | MAGNESIUM ION, N-[3-(4,5-diethyl-6-oxo-1,6-dihydropyrimidin-2-yl)-4-propoxyphenyl]-2-(4-methylpiperazin-1-yl)acetamide, ZINC ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Pharmacokinetics-Driven Optimization of 4(3 H)-Pyrimidinones as Phosphodiesterase Type 5 Inhibitors Leading to TPN171, a Clinical Candidate for the Treatment of Pulmonary Arterial Hypertension.
J.Med.Chem., 62, 2019
|
|
