7SVB
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite C | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
1XXH
 
 | | ATPgS Bound E. Coli Clamp Loader Complex | | Descriptor: | DNA polymerase III subunit gamma, DNA polymerase III, delta prime subunit, ... | | Authors: | Kazmirski, S.L, Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6IRQ
 
 | |
6IJU
 
 | |
8FII
 
 | | Wild type APOBEC3A in complex with TT(FdZ)-hairpin inhibitor (crystal form 1) | | Descriptor: | DNA (5'-D(P*GP*CP*GP*CP*TP*TP*(UFP)P*GP*CP*GP*C)-3'), DNA dC->dU-editing enzyme APOBEC-3A, PHOSPHATE ION, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | DNA-based inhibitors restrict mutagenic activity of APOBEC3 in cells
To Be Published
|
|
8FIJ
 
 | | Wild type APOBEC3A in complex with TT(FdZ)-hairpin inhibitor (crystal form 2) | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*TP*TP*(UFP)P*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, PHOSPHATE ION, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | DNA-based inhibitors restrict mutagenic activity of APOBEC3 in cells
To Be Published
|
|
6SJE
 
 | | Cryo-EM structure of the RecBCD Chi partially-recognised complex | | Descriptor: | DNA fork substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3SR2
 
 | | Crystal Structure of Human XLF-XRCC4 Complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Hammel, M, Classen, S, Tainer, J.A. | | Deposit date: | 2011-07-06 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.9708 Å) | | Cite: | XRCC4 Protein Interactions with XRCC4-like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair.
J.Biol.Chem., 286, 2011
|
|
8J70
 
 | | Native SAND domain of protein SP140 with DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*GP*CP*CP*GP*CP*CP*CP*T)-3'), Nuclear body protein SP140, selenourea | | Authors: | Li, H.T, Liu, W.Q. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for Speckled protein SP140 bivalent recognition of histone H3 and DNA.
To Be Published
|
|
6T7Y
 
 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T2V
 
 | | Cryo-EM structure of the RecBCD in complex with Chi-plus2 substrate | | Descriptor: | DNA (Chi-plus2), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6T7X
 
 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T2U
 
 | | Cryo-EM structure of the RecBCD in complex with Chi-minus2 substrate | | Descriptor: | DNA (Chi-minus2), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2VE8
 
 | |
5FF8
 
 | | TDG enzyme-product complex | | Descriptor: | DNA, G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
7RQT
 
 | | THE B-DNA DODECAMER With HIGH RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Chen, C, Huang, Z. | | Deposit date: | 2021-08-08 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | 2'-beta-Selenium Atom on Thymidine to Control beta-Form DNA Conformation and Large Crystal Formation
Cryst.Growth Des., 2022
|
|
5LKQ
 
 | | Protease domain of RadA | | Descriptor: | DNA repair protein RadA | | Authors: | Rapisarda, C, Fronzes, R. | | Deposit date: | 2016-07-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension.
Nat Commun, 8, 2017
|
|
327D
 
 | |
7Z13
 
 | | S. cerevisiae CMGE dimer nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
6B1R
 
 | |
6B1Q
 
 | |
5HF7
 
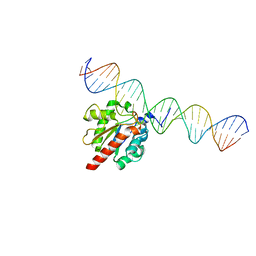 | | TDG enzyme-substrate complex | | Descriptor: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2016-01-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
6TUR
 
 | | human XPG, Apo1 form | | Descriptor: | DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
2R6C
 
 | | Crystal Form BH2 | | Descriptor: | DnaG Primase, Helicase Binding Domain, Replicative helicase | | Authors: | Bailey, S, Eliason, W.K, Steitz, T.A. | | Deposit date: | 2007-09-05 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase
Science, 318, 2007
|
|
2R6A
 
 | | Crystal Form BH1 | | Descriptor: | DnaG Primase, Helicase Binding Domain, Replicative helicase, ... | | Authors: | Bailey, S, Eliason, W.K, Steitz, T.A. | | Deposit date: | 2007-09-05 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase
Science, 318, 2007
|
|
