4TKJ
 
 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitic acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TLJ
 
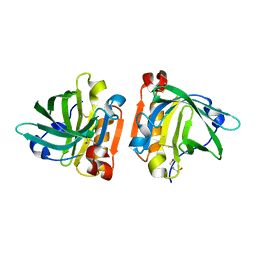 | | Ultra-high resolution crystal structure of caprine Beta-lactoglobulin | | Descriptor: | 1,4-BUTANEDIOL, Beta-lactoglobulin | | Authors: | Crowther, J.M, Jameson, G.B, Suzuki, H, Dobson, R.C.J. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ultra-high resolution crystal structure of recombinant caprine beta-lactoglobulin.
Febs Lett., 588, 2014
|
|
3FA6
 
 | |
3FEL
 
 | |
3FIQ
 
 | | Odorant Binding Protein OBP1 | | Descriptor: | 1,2-ETHANEDIOL, Odorant-binding protein 1F | | Authors: | White, S.A. | | Deposit date: | 2008-12-12 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structure of rat odorant-binding protein OBP1 at 1.6 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3FEP
 
 | | Crystal structure of the R132K:R111L:L121E:R59W-CRABPII mutant complexed with a synthetic ligand (merocyanin) at 2.60 angstrom resolution. | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-30 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
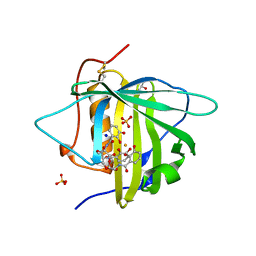
3CMP
 
 | |
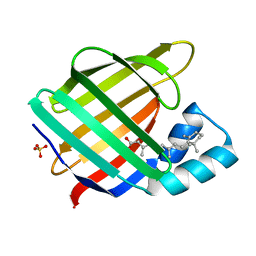
3CWK
 
 | |
3FA9
 
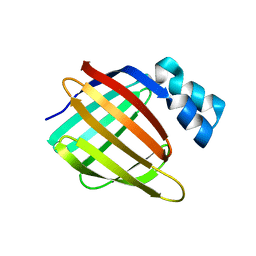 | |
3CR6
 
 | |
3D96
 
 | |
3DSZ
 
 | | Engineered human lipocalin 2 in complex with Y-DTPA | | Descriptor: | N-{(1S,2S)-2-[bis(carboxymethyl)amino]cyclohexyl}-N-{(2R)-2-[bis(carboxymethyl)amino]-3-[4-({[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]carbamothioyl}amino)phenyl]propyl}glycine, YTTRIUM (III) ION, engineered human lipocalin 2 | | Authors: | Eichinger, A, Skerra, A. | | Deposit date: | 2008-07-14 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-affinity recognition of lanthanide(III) chelate complexes by a reprogrammed human lipocalin 2
J.Am.Chem.Soc., 131, 2009
|
|
3FA8
 
 | |
3FEN
 
 | |
3DTQ
 
 | |
3EYC
 
 | | New crystal structure of human tear lipocalin in complex with 1,4-butanediol in space group P21 | | Descriptor: | 1,4-BUTANEDIOL, Lipocalin-1 | | Authors: | Breustedt, D.A, Keil, L, Skerra, A. | | Deposit date: | 2008-10-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new crystal form of human tear lipocalin reveals high flexibility in the loop region and induced fit in the ligand cavity
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3FMZ
 
 | |
3F9D
 
 | |
3FR5
 
 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 5-(3-carbamoylbenzyl)-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-4-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Uppenberg, J, Svensson, S, Rondahl, L, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FR2
 
 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 9-benzyl-2,3,4,9-tetrahydro-1H-carbazole-8-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Rondahl, L, Uppenberg, J, Svensson, S, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FA7
 
 | |
3FEK
 
 | |
3FW5
 
 | |
3FW4
 
 | |
3FR4
 
 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 9-[2-(trifluoromethyl)benzyl]-2,3,4,9-tetrahydro-1H-carbazole-8-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Uppenberg, J, Svensson, S, Rondahl, L, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
