1I6K
 
 | |
9G7I
 
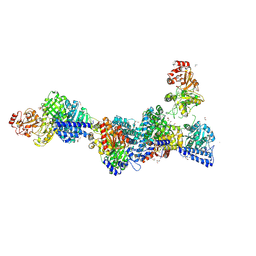 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with acetyl-Coenyzme A from Clostridium autoethanogenum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Lemaire, O.N, Yin, M.D, Murphy, B.J, Wagner, T. | | Deposit date: | 2024-07-21 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
9G8O
 
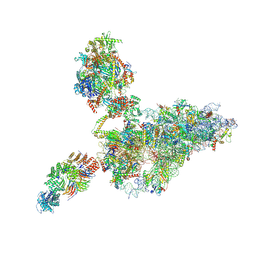 | | human 40S ribosome bound by a SKI238-exosome complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
1I3M
 
 | | MOLECULAR BASIS FOR SEVERE EPIMERASE-DEFICIENCY GALACTOSEMIA: X-RAY STRUCTURE OF THE HUMAN V94M-SUBSTITUTED UDP-GALACTOSE 4-EPIMERASE | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2001-02-15 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase.
J.Biol.Chem., 276, 2001
|
|
1I6L
 
 | |
9G8M
 
 | | human 80S ribosome bound by a SKI2-exosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
1I3K
 
 | | MOLECULAR BASIS FOR SEVERE EPIMERASE-DEFICIENCY GALACTOSEMIA: X-RAY STRUCTURE OF THE HUMAN V94M-SUBSTITUTED UDP-GALACTOSE 4-EPIMERASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2001-02-15 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase.
J.Biol.Chem., 276, 2001
|
|
9ETM
 
 | |
5D5O
 
 | | HcgC from Methanocaldococcus jannaschii | | Descriptor: | SULFATE ION, Uncharacterized protein MJ0489 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8QUP
 
 | |
1IQX
 
 | | CRYSTAL STRUCTURE OF COBALT-SUBSTITUTED AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS | | Descriptor: | CO(II)-SUBSTITUTED AMINE OXIDASE, COBALT (II) ION | | Authors: | Kishishita, S, Okajima, T, Mure, M, Kim, M, Yamaguchi, H, Hirota, S, Suzuki, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2001-08-27 | | Release date: | 2003-02-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Copper Ion in Bacterial Copper Amine Oxidase: Spectroscopic and Crystallographic Studies of Metal-Substituted Enzymes
J.AM.CHEM.SOC., 125, 2003
|
|
1IU7
 
 | | HOLO FORM OF COPPER-CONTAINING AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS | | Descriptor: | AMINE OXIDASE, COPPER (II) ION | | Authors: | Kishishita, S, Okajima, T, Kim, M, Yamaguchi, H, Hirota, S, Suzuki, S, Kuroda, S, Tanizawa, K, Mure, M. | | Deposit date: | 2002-02-28 | | Release date: | 2003-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Copper Ion in Bacterial Copper Amine Oxidase: Spectroscopic and Crystallographic Studies of Metal-Substituted Enzymes
J.AM.CHEM.SOC., 125, 2003
|
|
1IW8
 
 | | Crystal Structure of a mutant of acid phosphatase from Escherichia blattae (G74D/I153T) | | Descriptor: | SULFATE ION, acid phosphatase | | Authors: | Ishikawa, K, Mihara, Y, Shimba, N, Ohtsu, N, Kawasaki, H, Suzuki, E, Asano, Y. | | Deposit date: | 2002-04-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhancement of nucleoside phosphorylation activity in an acid phosphatase
PROTEIN ENG., 15, 2002
|
|
3CR4
 
 | | X-ray structure of bovine Pnt,Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3CL0
 
 | | N1 Neuraminidase H274Y + oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Collins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
3CIV
 
 | |
1MDY
 
 | | CRYSTAL STRUCTURE OF MYOD BHLH DOMAIN BOUND TO DNA: PERSPECTIVES ON DNA RECOGNITION AND IMPLICATIONS FOR TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*CP*AP*GP*CP*TP*GP*TP*TP*GP*A)-3'), PROTEIN (MYOD BHLH DOMAIN) | | Authors: | Ma, P.C.M, Rould, M.A, Weintraub, H, Pabo, C.O. | | Deposit date: | 1994-06-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation.
Cell(Cambridge,Mass.), 77, 1994
|
|
3CO0
 
 | |
3DEN
 
 | | Structure of E. coli DHDPS mutant Y107W | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Pearce, F.G, Gerrard, J.A, Perugini, M.A, Jameson, G.B. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutating the tight-dimer interface of dihydrodipicolinate synthase disrupts the enzyme quaternary structure: toward a monomeric enzyme
Biochemistry, 47, 2008
|
|
3DFJ
 
 | | Crystal structure of human Prostasin | | Descriptor: | CHLORIDE ION, Prostasin | | Authors: | Su, H.P, Rickert, K.W, Darke, P.L, Munshi, S.K. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of human prostasin, a target for the regulation of hypertension.
J.Biol.Chem., 283, 2008
|
|
3DFL
 
 | | Crystal structure of human Prostasin complexed to 4-guanidinobenzoic acid | | Descriptor: | 4-carbamimidamidobenzoic acid, Prostasin | | Authors: | Su, H.P, Rickert, K.W, Darke, P.L, Munshi, S.K. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human prostasin, a target for the regulation of hypertension.
J.Biol.Chem., 283, 2008
|
|
3D8X
 
 | | Crystal Structure of Saccharomyces cerevisiae NDPPH Dependent Thioredoxin Reductase 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1 | | Authors: | Zhang, Z.Y, Bao, R, Yu, J, Chen, Y.X, Zhou, C.-Z. | | Deposit date: | 2008-05-26 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae cytoplasmic thioredoxin reductase Trr1 reveals the structural basis for species-specific recognition of thioredoxin
Biochim.Biophys.Acta, 1794, 2009
|
|
1S7F
 
 | | RimL- Ribosomal L7/L12 alpha-N-protein acetyltransferase crystal form I (apo) | | Descriptor: | CHLORIDE ION, MALONIC ACID, acetyl transferase | | Authors: | Vetting, M.W, de Carvalho, L.P, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium.
J.Biol.Chem., 280, 2005
|
|
2JQH
 
 | | VPS4B MIT | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2K0M
 
 | | Solution NMR structure of the uncharacterized protein from Rhodospirillum rubrum gene locus Rru_A0810. Northeast Structural Genomics Target RrR43 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Wang, H, Jiang, M, Foote, E.L, Xiao, R, Liu, J, Swapna, G, Acton, T.B, Baran, M.C, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the uncharacterized protein from Rhodospirillum rubrum gene locus Rru_A0810. Northeast Structural Genomics Target RrR43.
To be Published
|
|
