3UEF
 
 | | Crystal structure of human Survivin bound to histone H3 (C2 space group). | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3NX4
 
 | | Crystal structure of the yhdH oxidoreductase from Salmonella enterica in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Peterson, S, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-12 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the yhdH oxidoreductase from Salmonella enterica in complex with NADP
To be Published
|
|
3O04
 
 | | Crystal structure of the beta-keto-acyl carrier protein synthase II (lmo2201) from Listeria monocytogenes | | Descriptor: | beta-keto-acyl carrier protein synthase II | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the beta-keto-acyl carrier protein synthase II (lmo2201) from Listeria monocytogenes
To be Published
|
|
3UEG
 
 | | Crystal structure of human Survivin K62A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, TETRAETHYLENE GLYCOL, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEI
 
 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3WR1
 
 | | Crystal structure of Cormorant (Phalacrocorax carbo) hemoglobin | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jagadeesan, G, Vinodh Kumar, V, Peters, H.G, Malathy, P, Harikrishna Etti, S, Gunasekaran, K, Aravindhan, S. | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of Cormorant (Phalacrocorax carbo) hemoglobin
To be Published
|
|
4U6N
 
 | | Crystal structure of Escherichia coli DiaA | | Descriptor: | CHLORIDE ION, DnaA initiator-associating protein DiaA | | Authors: | Oakley, A.J, Lo, T. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Escherichia coli DiaA
To Be Published
|
|
3TKD
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and cyclothiazide at 1.45 A resolution | | Descriptor: | CYCLOTHIAZIDE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Krintel, C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-08-26 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thermodynamics and structural analysis of positive allosteric modulation of the ionotropic glutamate receptor GluA2.
Biochem.J., 441, 2012
|
|
3U8K
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3573 (1-(5-ethoxypyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
5OGW
 
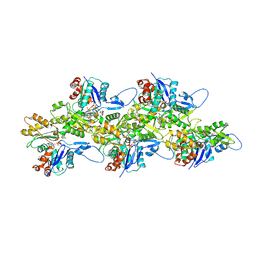 | | Cryo-EM structure of jasplakinolide-stabilized malaria parasite F-actin at near-atomic resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, Jasplakinolide, ... | | Authors: | Pospich, S, Kumpula, E.-P, von der Ecken, J, Vahokoski, J, Kursula, I, Raunser, S. | | Deposit date: | 2017-07-13 | | Release date: | 2017-09-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Near-atomic structure of jasplakinolide-stabilized malaria parasite F-actin reveals the structural basis of filament instability.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3IN8
 
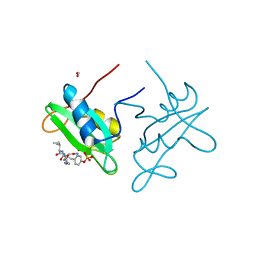 | |
3IMJ
 
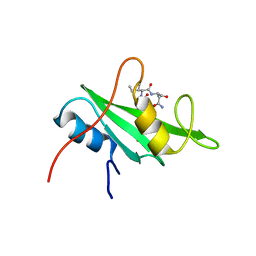 | |
3SWP
 
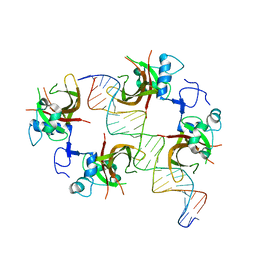 | | ANAC019 NAC domain in complex with DNA | | Descriptor: | NAC domain-containing protein 19, oligonucleotide forward, oligonucleotide reverse | | Authors: | Welner, D, Lo Leggio, L. | | Deposit date: | 2011-07-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.113 Å) | | Cite: | DNA binding by the plant-specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors.
Biochem.J., 444, 2012
|
|
8OID
 
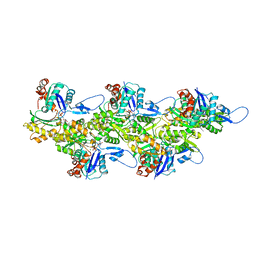 | | Cryo-EM structure of ADP-bound, filamentous beta-actin harboring the N111S mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OI6
 
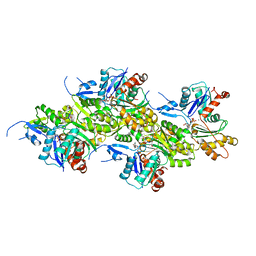 | | Cryo-EM structure of the undecorated barbed end of filamentous beta/gamma actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OI8
 
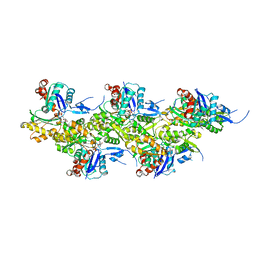 | | Cryo-EM structure of ADP-bound, filamentous beta-actin harboring the R183W mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3V0L
 
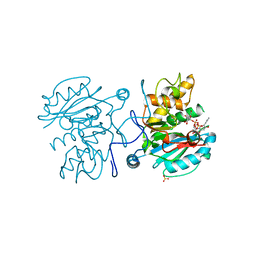 | | Crystal structure of the Fucosylgalactoside alpha N-acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with a novel UDP-Gal derived inhibitor (2GW) | | Descriptor: | 5-phenyl-uridine-5'-alpha-d-galactosyl-diphosphate, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Palcic, M.M, Jorgensen, R. | | Deposit date: | 2011-12-08 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Base-modified Donor Analogues Reveal Novel Dynamic Features of a Glycosyltransferase.
J.Biol.Chem., 288, 2013
|
|
8OIE
 
 | | Iron Nitrogenase Complex from Rhodobacter capsulatus | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Schmidt, F.V, Schulz, L, Zarzycki, J, Prinz, S, Erb, T.J, Rebelein, J.G. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural insights into the iron nitrogenase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OUA
 
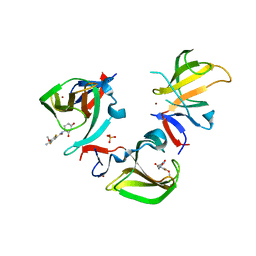 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11f | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2,5-bis(fluoranyl)-3-methoxy-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-22 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU4
 
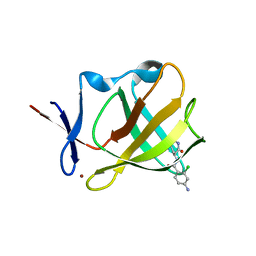 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11a | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-chloranyl-benzamide, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU6
 
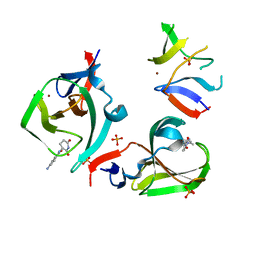 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11c | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-methyl-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU3
 
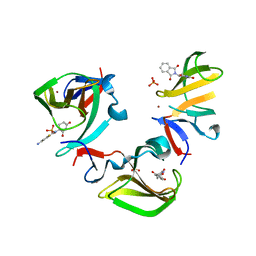 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 8d | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-fluoranyl-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Heim, C, Bischof, L, Maiwald, S, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU5
 
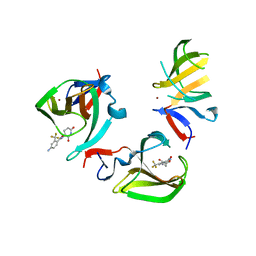 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11b | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-(trifluoromethyl)benzamide, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU7
 
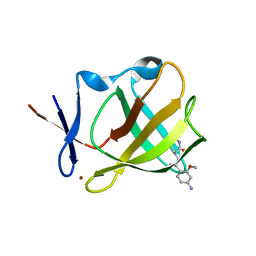 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11d | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-methoxy-benzamide, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-22 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU9
 
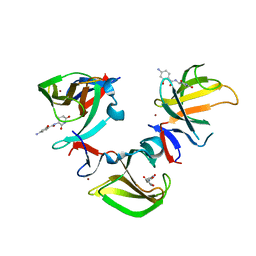 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11e | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-oxidanyl-benzamide, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-22 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
