1NDQ
 
 | | Bacillus lentus subtilisin | | Descriptor: | CALCIUM ION, Subtilisin Savinase | | Authors: | Pan, X, Bott, R, Glatz, C.E. | | Deposit date: | 2002-12-09 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Subtilisin surface properties and crystal growth kinetics
J.CRYST.GROWTH, 254, 2003
|
|
7GSL
 
 | |
1NFD
 
 | | AN ALPHA-BETA T CELL RECEPTOR (TCR) HETERODIMER IN COMPLEX WITH AN ANTI-TCR FAB FRAGMENT DERIVED FROM A MITOGENIC ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H57 FAB, N15 ALPHA-BETA T-CELL RECEPTOR | | Authors: | Wang, J.-H, Lim, K, Smolyar, A, Teng, M.-K, Sacchittini, J, Reinherz, E.L. | | Deposit date: | 1997-08-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody.
EMBO J., 17, 1998
|
|
1SL7
 
 | | Crystal structure of calcium-loaded apo-obelin from Obelia longissima | | Descriptor: | CALCIUM ION, Obelin | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
7JSJ
 
 | | Structure of the NaCT-PF2 complex | | Descriptor: | (2R)-2-[2-(4-tert-butylphenyl)ethyl]-2-hydroxybutanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Sauer, D.B, Wang, B, Song, J, Rice, W.J, Wang, D.N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure and inhibition mechanism of the human citrate transporter NaCT.
Nature, 591, 2021
|
|
7JMR
 
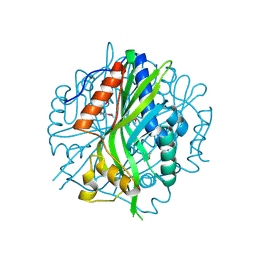 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pea pathogenicity protein 2 | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
7JMV
 
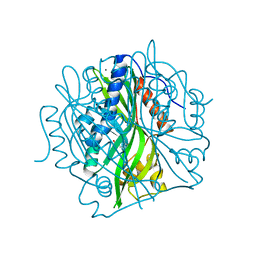 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis complexed with 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
7K15
 
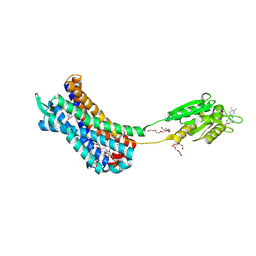 | | Crystal structure of the Human Leukotriene B4 Receptor 1 in Complex with Selective Antagonist MK-D-046 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLAVIN MONONUCLEOTIDE, HEXAETHYLENE GLYCOL, ... | | Authors: | Michaelian, N, Han, G.W, Cherezov, V. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights on ligand recognition at the human leukotriene B4 receptor 1.
Nat Commun, 12, 2021
|
|
1C53
 
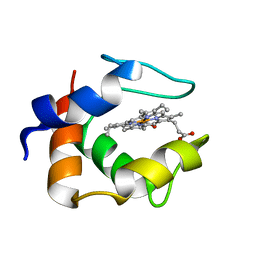 | | S-CLASS CYTOCHROMES C HAVE A VARIETY OF FOLDING PATTERNS: STRUCTURE OF CYTOCHROME C-553 FROM DESULFOVIBRIO VULGARIS DETERMINED BY THE MULTI-WAVELENGTH ANOMALOUS DISPERSION METHOD | | Descriptor: | CYTOCHROME C553, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakagawa, A, Higuchi, Y, Yasuoka, N, Katsube, Y, Yaga, T. | | Deposit date: | 1991-08-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-class cytochromes c have a variety of folding patterns: structure of cytochrome c-553 from Desulfovibrio vulgaris determined by the multi-wavelength anomalous dispersion method.
J.Biochem.(Tokyo), 108, 1990
|
|
1SKU
 
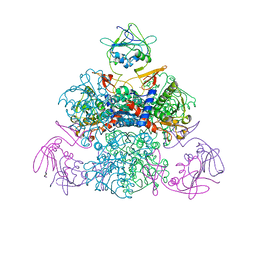 | | E. coli Aspartate Transcarbamylase 240's Loop Mutant (K244N) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, MALONATE ION, ... | | Authors: | Alam, N, Stieglitz, K.A, Caban, M.D, Gourinath, S, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 240s Loop Interactions Stabilize the T State of Escherichia coli Aspartate Transcarbamoylase.
J.Biol.Chem., 279, 2004
|
|
7JI3
 
 | |
1NAW
 
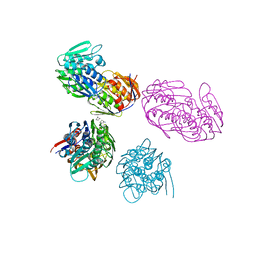 | | ENOLPYRUVYL TRANSFERASE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | Authors: | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | Deposit date: | 1996-07-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
1Y39
 
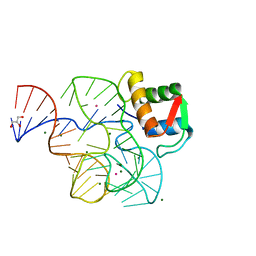 | | Co-evolution of protein and RNA structures within a highly conserved ribosomal domain | | Descriptor: | 50S ribosomal protein L11, 58 Nucleotide Ribosomal 23S RNA Domain, COBALT (III) ION, ... | | Authors: | Dunstan, M.S, GuhaThakurta, D, Draper, D.E, Conn, G.L. | | Deposit date: | 2004-11-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coevolution of Protein and RNA Structures within a Highly Conserved Ribosomal Domain
Chem.Biol., 12, 2005
|
|
7JSK
 
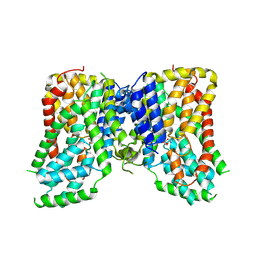 | | Structure of the NaCT-Citrate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, SODIUM ION, ... | | Authors: | Sauer, D.B, Wang, B, Song, J, Rice, W.J, Wang, D.N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure and inhibition mechanism of the human citrate transporter NaCT.
Nature, 591, 2021
|
|
7JMN
 
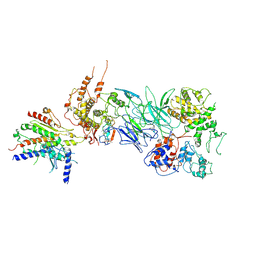 | | Tail module of Mediator complex | | Descriptor: | MED15, Mediator of RNA polymerase II transcription subunit 14, Mediator of RNA polymerase II transcription subunit 16, ... | | Authors: | Zhang, H.Q, Chen, D.C. | | Deposit date: | 2020-08-02 | | Release date: | 2021-03-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
7JVR
 
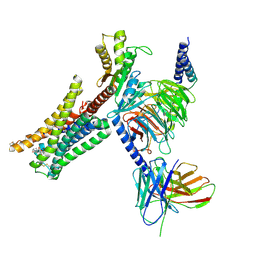 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
1NG0
 
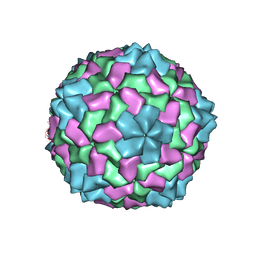 | |
7JWR
 
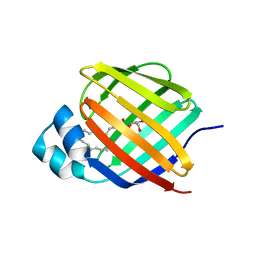 | |
7JX2
 
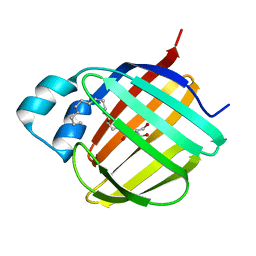 | |
1T34
 
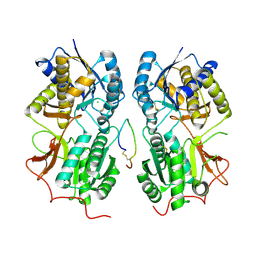 | | ROTATION MECHANISM FOR TRANSMEMBRANE SIGNALING BY THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Atrial natriuretic peptide factor, Atrial natriuretic peptide receptor A, ... | | Authors: | Ogawa, H, Qiu, Y, Ogata, C.M, Misono, K.S. | | Deposit date: | 2004-04-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction
J.Biol.Chem., 279, 2004
|
|
7JVG
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 1-arachidonoylglycerol | | Descriptor: | (2S)-2,3-dihydroxypropyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Golczak, M. | | Deposit date: | 2020-08-21 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
7JWD
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 2-linoleoylglycerol | | Descriptor: | 1,3-dihydroxypropan-2-yl (9Z,12Z)-octadeca-9,12-dienoate, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Banarjee, S, Golczak, M. | | Deposit date: | 2020-08-25 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35000193 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
1T4M
 
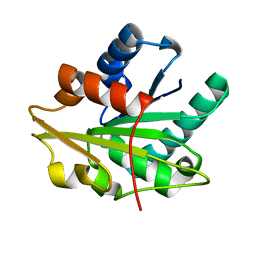 | |
7JVY
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 2-arachidonylglyceryl ether | | Descriptor: | 2-{[(5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraen-1-yl]oxy}propane-1,3-diol, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Banarjee, S, Golczak, M. | | Deposit date: | 2020-08-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
7K3I
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 2-lauroylglycerol | | Descriptor: | 1,3-dihydroxypropan-2-yl dodecanoate, Retinol-binding protein 2 | | Authors: | Adams, C, Silvaroli, J.A, Banarjee, S, Golczak, M. | | Deposit date: | 2020-09-11 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
