7JMX
 
 | | Crystal structure of a SARS-CoV-2 cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | ACETATE ION, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
7JV2
 
 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody Fab fragment (local refinement of the receptor-binding motif and Fab variable domains) | | Descriptor: | S2H13 Fab heavy chain, S2H13 Fab light chain, Spike glycoprotein | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JW0
 
 | | SARS-CoV-2 spike in complex with the S304 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S304 Fab heavy chain, ... | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7LKA
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 | | Descriptor: | ACETATE ION, COV107-23 heavy chain, COV107-23 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
7LK9
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 HC + COVD21-C8 LC | | Descriptor: | COV107-23 heavy chain, COVD21-C8 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-01 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
7LOP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV05-163 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab heavy chain, CR3022 Fab light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-10 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants.
Science, 373, 2021
|
|
7JTL
 
 | | Structure of SARS-CoV-2 ORF8 accessory protein | | Descriptor: | ORF8 protein, SODIUM ION | | Authors: | Flower, T.G, Buffalo, C.Z, Hooy, R.M, Allaire, M, Ren, X, Hurley, J.H. | | Deposit date: | 2020-08-18 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JVZ
 
 | |
7JVA
 
 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment (local refinement of the receptor-binding domain and Fab variable domains) | | Descriptor: | S2A4 Fab heavy chain, S2A4 Fab light chain, Spike glycoprotein, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7M02
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 17c | | Descriptor: | (1R,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZY
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M01
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZT
 
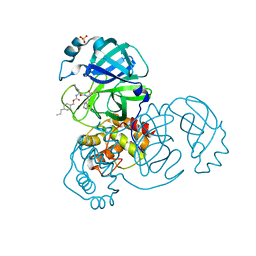 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8b | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZX
 
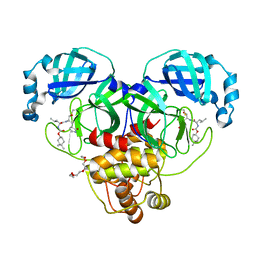 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 1c | | Descriptor: | (1R,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZU
 
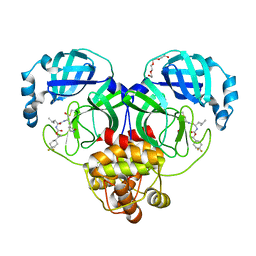 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 12b | | Descriptor: | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M04
 
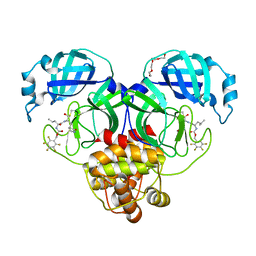 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 21c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZW
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 20b (deuterated analog of 19b) | | Descriptor: | (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZZ
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 5c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1r,4S)-4-phenylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1r,4S)-4-phenylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M03
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 18c | | Descriptor: | (1R,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZV
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 19b | | Descriptor: | (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M00
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 13c | | Descriptor: | (1R,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LCQ
 
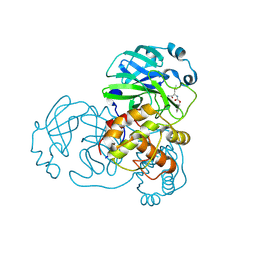 | | N-terminal finger stabilizes feline drug GC376 in coronavirus 3CL protease | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-Terminal Finger Stabilizes the S1 Pocket for the Reversible Feline Drug GC376 in the SARS-CoV-2 M pro Dimer.
J.Mol.Biol., 433, 2021
|
|
7LCP
 
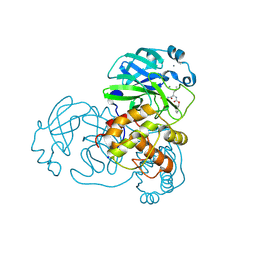 | | N-terminal finger stabilizes feline drug GC376 in coronavirus 3CL protease | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Khan, M.B, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N-Terminal Finger Stabilizes the S1 Pocket for the Reversible Feline Drug GC376 in the SARS-CoV-2 M pro Dimer.
J.Mol.Biol., 433, 2021
|
|
7L8I
 
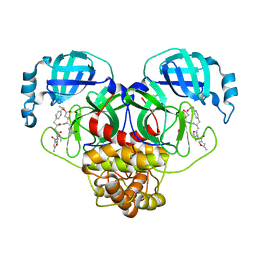 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21) | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
7L8J
 
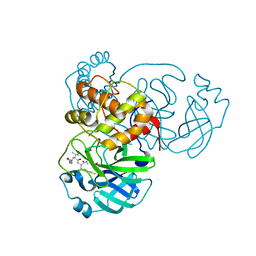 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21212) | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
